Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity
- PMID: 25313155
- PMCID: PMC4227783
- DOI: 10.1093/nar/gku942
Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity
Abstract
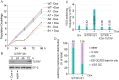
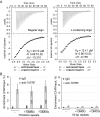
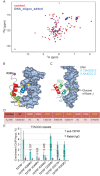
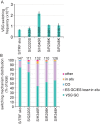
Trypanosoma brucei causes human African trypanosomiasis and regularly switches its major surface antigen, VSG, in the bloodstream of its mammalian host to evade the host immune response. VSGs are expressed exclusively from subtelomeric loci, and we have previously shown that telomere proteins TbTIF2 and TbRAP1 play important roles in VSG switching and VSG silencing regulation, respectively. We now discover that the telomere duplex DNA-binding factor, TbTRF, also plays a critical role in VSG switching regulation, as a transient depletion of TbTRF leads to significantly more VSG switching events. We solved the NMR structure of the DNA-binding Myb domain of TbTRF, which folds into a canonical helix-loop-helix structure that is conserved to the Myb domains of mammalian TRF proteins. The TbTRF Myb domain tolerates well the bulky J base in T. brucei telomere DNA, and the DNA-binding affinity of TbTRF is not affected by the presence of J both in vitro and in vivo. In addition, we find that point mutations in TbTRF Myb that significantly reduced its in vivo telomere DNA-binding affinity also led to significantly increased VSG switching frequencies, indicating that the telomere DNA-binding activity is critical for TbTRF's role in VSG switching regulation.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions.mSphere. 2020 Feb 26;5(1):e00027-20. doi: 10.1128/mSphere.00027-20. mSphere. 2020. PMID: 32102938 Free PMC article.
-
Trypanosoma brucei TIF2 and TRF Suppress VSG Switching Using Overlapping and Independent Mechanisms.PLoS One. 2016 Jun 3;11(6):e0156746. doi: 10.1371/journal.pone.0156746. eCollection 2016. PLoS One. 2016. PMID: 27258069 Free PMC article.
-
TbTRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain.Nucleic Acids Res. 2021 Jun 4;49(10):5637-5653. doi: 10.1093/nar/gkab401. Nucleic Acids Res. 2021. PMID: 34048580 Free PMC article.
-
Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities.Pathogens. 2021 Jul 30;10(8):967. doi: 10.3390/pathogens10080967. Pathogens. 2021. PMID: 34451431 Free PMC article. Review.
-
Telomere and Subtelomere R-loops and Antigenic Variation in Trypanosomes.J Mol Biol. 2020 Jul 10;432(15):4167-4185. doi: 10.1016/j.jmb.2019.10.025. Epub 2019 Nov 2. J Mol Biol. 2020. PMID: 31682833 Free PMC article. Review.
Cited by
-
Editorial: Telomere Flexibility and Versatility: A Role of Telomeres in Adaptive Potential.Front Genet. 2021 Oct 4;12:771938. doi: 10.3389/fgene.2021.771938. eCollection 2021. Front Genet. 2021. PMID: 34671387 Free PMC article. No abstract available.
-
Epigenetic Regulation of Virulence Gene Expression in Parasitic Protozoa.Cell Host Microbe. 2016 May 11;19(5):629-40. doi: 10.1016/j.chom.2016.04.020. Cell Host Microbe. 2016. PMID: 27173931 Free PMC article. Review.
-
TbRAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing.Sci Adv. 2020 Sep 18;6(38):eabc4065. doi: 10.1126/sciadv.abc4065. Print 2020 Sep. Sci Adv. 2020. PMID: 32948591 Free PMC article.
-
How to create coats for all seasons: elucidating antigenic variation in African trypanosomes.Emerg Top Life Sci. 2017 Dec 22;1(6):593-600. doi: 10.1042/ETLS20170105. Emerg Top Life Sci. 2017. PMID: 33525853 Free PMC article.
-
Genome maintenance functions of a putative Trypanosoma brucei translesion DNA polymerase include telomere association and a role in antigenic variation.Nucleic Acids Res. 2020 Sep 25;48(17):9660-9680. doi: 10.1093/nar/gkaa686. Nucleic Acids Res. 2020. PMID: 32890403 Free PMC article.
References
-
- Barry J.D., McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. - PubMed
-
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. - PubMed
-
- Cross G.A.M., Kim H.S., Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 2014;195:59–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

