Characterization of a novel plasmid-borne thiopeptide gene cluster in Staphylococcus epidermidis strain 115
- PMID: 25313391
- PMCID: PMC4248843
- DOI: 10.1128/JB.02243-14
Characterization of a novel plasmid-borne thiopeptide gene cluster in Staphylococcus epidermidis strain 115
Abstract
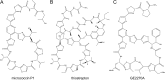
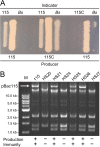
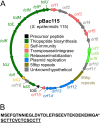
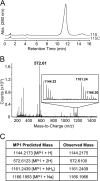
Thiopeptides are small (12- to 17-amino-acid), heavily modified peptides of bacterial origin. This antibiotic family, with more than 100 known members, is characterized by the presence of sulfur-containing heterocyclic rings and dehydrated residues within a macrocyclic peptide structure. Thiopeptides, including micrococcin P1, have garnered significant attention in recent years for their potent antimicrobial activity against bacteria, fungi, and even protozoa. Micrococcin P1 is known to target the ribosome; however, like those of other thiopeptides, its biosynthesis and mechanisms of self-immunity are poorly characterized. We have discovered an isolate of Staphylococcus epidermidis harboring the genes for thiopeptide production and self-protection on a 24-kb plasmid. Here we report the characterization of this plasmid, identify the antimicrobial peptide that it encodes, and provide evidence of a target replacement-mediated mechanism of self-immunity.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Elucidating and engineering thiopeptide biosynthesis.World J Microbiol Biotechnol. 2017 Jun;33(6):119. doi: 10.1007/s11274-017-2283-9. Epub 2017 May 11. World J Microbiol Biotechnol. 2017. PMID: 28497389 Review.
-
ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences.PLoS One. 2012;7(9):e45878. doi: 10.1371/journal.pone.0045878. Epub 2012 Sep 24. PLoS One. 2012. PMID: 23029291 Free PMC article.
-
Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci.Antimicrob Agents Chemother. 2012 Mar;56(3):1539-47. doi: 10.1128/AAC.05397-11. Epub 2011 Dec 12. Antimicrob Agents Chemother. 2012. PMID: 22155816 Free PMC article.
-
Reconstitution and Minimization of a Micrococcin Biosynthetic Pathway in Bacillus subtilis.J Bacteriol. 2016 Aug 25;198(18):2431-8. doi: 10.1128/JB.00396-16. Print 2016 Sep 15. J Bacteriol. 2016. PMID: 27381911 Free PMC article.
-
Chemical optimization and derivatization of micrococcin p2 to target multiple bacterial infections: new antibiotics from thiopeptides.World J Microbiol Biotechnol. 2024 Aug 20;40(10):307. doi: 10.1007/s11274-024-04109-5. World J Microbiol Biotechnol. 2024. PMID: 39162916 Review.
Cited by
-
Elucidating and engineering thiopeptide biosynthesis.World J Microbiol Biotechnol. 2017 Jun;33(6):119. doi: 10.1007/s11274-017-2283-9. Epub 2017 May 11. World J Microbiol Biotechnol. 2017. PMID: 28497389 Review.
-
Catch me if you can: capturing microbial community transformation by extracellular DNA using Hi-C sequencing.Antonie Van Leeuwenhoek. 2023 Jul;116(7):667-685. doi: 10.1007/s10482-023-01834-z. Epub 2023 May 8. Antonie Van Leeuwenhoek. 2023. PMID: 37156983 Free PMC article.
-
Genomic Analysis of Bacteriocin-Producing Staphylococci: High Prevalence of Lanthipeptides and the Micrococcin P1 Biosynthetic Gene Clusters.Probiotics Antimicrob Proteins. 2025 Feb;17(1):159-174. doi: 10.1007/s12602-023-10119-w. Epub 2023 Aug 26. Probiotics Antimicrob Proteins. 2025. PMID: 37632676 Free PMC article.
-
Antimicrobials from a feline commensal bacterium inhibit skin infection by drug-resistant S. pseudintermedius.Elife. 2021 Oct 19;10:e66793. doi: 10.7554/eLife.66793. Elife. 2021. PMID: 34664551 Free PMC article.
-
A Strong Synergy Between the Thiopeptide Bacteriocin Micrococcin P1 and Rifampicin Against MRSA in a Murine Skin Infection Model.Front Immunol. 2021 Jul 2;12:676534. doi: 10.3389/fimmu.2021.676534. eCollection 2021. Front Immunol. 2021. PMID: 34276663 Free PMC article.
References
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30:108–160. 10.1039/c2np20085f. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

