Integrated mRNA-microRNA profiling of human NK cell differentiation identifies MiR-583 as a negative regulator of IL2Rγ expression
- PMID: 25313504
- PMCID: PMC4196775
- DOI: 10.1371/journal.pone.0108913
Integrated mRNA-microRNA profiling of human NK cell differentiation identifies MiR-583 as a negative regulator of IL2Rγ expression
Abstract
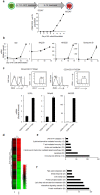
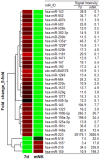
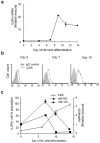
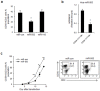
Natural killer (NK) cells are innate immune effector cells that protect against cancer and some viral infections. Until recently, most studies have investigated the molecular signatures of human or mouse NK cells to identify genes that are specifically expressed during NK cell development. However, the mechanism regulating NK cell development remains unclear. Here, we report a regulatory network of potential interactions during in vitro differentiation of human NK cells, identified using genome-wide mRNA and miRNA databases through hierarchical clustering analysis, gene ontology analysis and a miRNA target prediction program. The microRNA (miR)-583, which demonstrated the largest ratio change in mature NK cells, was highly correlated with IL2 receptor gamma (IL2Rγ) expression. The overexpression of miR-583 had an inhibitory effect on NK cell differentiation. In a reporter assay, the suppressive effect of miR-583 was ablated by mutating the putative miR-583 binding site of the IL2Rγ 3' UTR. Therefore, we show that miR-583 acts as a negative regulator of NK cell differentiation by silencing IL2Rγ. Additionally, we provide a comprehensive database of genome-wide mRNA and miRNA expression during human NK cell differentiation, offering a better understanding of basic human NK cell biology for the application of human NK cells in immunotherapy.
Conflict of interest statement
Figures


References
-
- Di Santo JP (2006) Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol 24: 257–286. - PubMed
-
- Galy A, Travis M, Cen D, Chen B (1995) Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 3: 459–473. - PubMed
-
- Blom B, Spits H (2006) Development of human lymphoid cells. Annu Rev Immunol 24: 287–320. - PubMed
-
- Boggs SS, Trevisan M, Patrene K, Geogopoulos K (1998) Lack of natural killer cell precursors in fetal liver of Ikaros knockout mutant mice. Nat Immun 16: 137–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

