Covariance of charged amino acids at positions 322 and 440 of HIV-1 Env contributes to coreceptor specificity of subtype B viruses, and can be used to improve the performance of V3 sequence-based coreceptor usage prediction algorithms
- PMID: 25313689
- PMCID: PMC4196930
- DOI: 10.1371/journal.pone.0109771
Covariance of charged amino acids at positions 322 and 440 of HIV-1 Env contributes to coreceptor specificity of subtype B viruses, and can be used to improve the performance of V3 sequence-based coreceptor usage prediction algorithms
Abstract
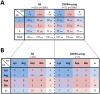
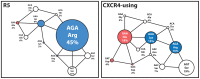
The ability to determine coreceptor usage of patient-derived human immunodeficiency virus type 1 (HIV-1) strains is clinically important, particularly for the administration of the CCR5 antagonist maraviroc. The envelope glycoprotein (Env) determinants of coreceptor specificity lie primarily within the gp120 V3 loop region, although other Env determinants have been shown to influence gp120-coreceptor interactions. Here, we determined whether conserved amino acid alterations outside the V3 loop that contribute to coreceptor usage exist, and whether these alterations improve the performance of V3 sequence-based coreceptor usage prediction algorithms. We demonstrate a significant covariant association between charged amino acids at position 322 in V3 and position 440 in the C4 Env region that contributes to the specificity of HIV-1 subtype B strains for CCR5 or CXCR4. Specifically, positively charged Lys/Arg at position 322 and negatively charged Asp/Glu at position 440 occurred more frequently in CXCR4-using viruses, whereas negatively charged Asp/Glu at position 322 and positively charged Arg at position 440 occurred more frequently in R5 strains. In the context of CD4-bound gp120, structural models suggest that covariation of amino acids at Env positions 322 and 440 has the potential to alter electrostatic interactions that are formed between gp120 and charged amino acids in the CCR5 N-terminus. We further demonstrate that inclusion of a "440 rule" can improve the sensitivity of several V3 sequence-based genotypic algorithms for predicting coreceptor usage of subtype B HIV-1 strains, without compromising specificity, and significantly improves the AUROC of the geno2pheno algorithm when set to its recommended false positive rate of 5.75%. Together, our results provide further mechanistic insights into the intra-molecular interactions within Env that contribute to coreceptor specificity of subtype B HIV-1 strains, and demonstrate that incorporation of Env determinants outside V3 can improve the reliability of coreceptor usage prediction algorithms.
Conflict of interest statement
Figures

Similar articles
-
Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains.J Virol. 2005 Jun;79(11):6957-68. doi: 10.1128/JVI.79.11.6957-6968.2005. J Virol. 2005. PMID: 15890935 Free PMC article.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
-
Hybrid approach for predicting coreceptor used by HIV-1 from its V3 loop amino acid sequence.PLoS One. 2013 Apr 15;8(4):e61437. doi: 10.1371/journal.pone.0061437. Print 2013. PLoS One. 2013. PMID: 23596523 Free PMC article.
-
Role of naturally occurring basic amino acid substitutions in the human immunodeficiency virus type 1 subtype E envelope V3 loop on viral coreceptor usage and cell tropism.J Virol. 1999 Jul;73(7):5520-6. doi: 10.1128/JVI.73.7.5520-5526.1999. J Virol. 1999. PMID: 10364300 Free PMC article.
-
HIV-1 tropism determination using a phenotypic Env recombinant viral assay highlights overestimation of CXCR4-usage by genotypic prediction algorithms for CRF01_AE and CRF02_AG [corrected].PLoS One. 2013 May 8;8(5):e60566. doi: 10.1371/journal.pone.0060566. Print 2013. PLoS One. 2013. PMID: 23667426 Free PMC article.
Cited by
-
Substitution of gp120 C4 region compensates for V3 loss-of-fitness mutations in HIV-1 CRF01_AE co-receptor switching.Emerg Microbes Infect. 2023 Dec;12(1):e2169196. doi: 10.1080/22221751.2023.2169196. Emerg Microbes Infect. 2023. PMID: 36647730 Free PMC article.
-
Evolution of coreceptor utilization to escape CCR5 antagonist therapy.Virology. 2016 Jul;494:198-214. doi: 10.1016/j.virol.2016.04.010. Epub 2016 Apr 26. Virology. 2016. PMID: 27128349 Free PMC article.
-
Individuals with HIV-1 Subtype C Infection and Cryptococcal Meningitis Exhibit Viral Genetic Intermixing of HIV-1 Between Plasma and Cerebrospinal Fluid and a High Prevalence of CXCR4-Using Variants.AIDS Res Hum Retroviruses. 2018 Jul;34(7):607-620. doi: 10.1089/AID.2017.0209. Epub 2018 May 23. AIDS Res Hum Retroviruses. 2018. PMID: 29658309 Free PMC article.
-
Maternal Binding and Neutralizing IgG Responses Targeting the C-Terminal Region of the V3 Loop Are Predictive of Reduced Peripartum HIV-1 Transmission Risk.J Virol. 2017 Apr 13;91(9):e02422-16. doi: 10.1128/JVI.02422-16. Print 2017 May 1. J Virol. 2017. PMID: 28202762 Free PMC article.
-
Reliable genotypic tropism tests for the major HIV-1 subtypes.Sci Rep. 2015 Feb 25;5:8543. doi: 10.1038/srep08543. Sci Rep. 2015. PMID: 25712827 Free PMC article.
References
-
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, et al. (1996) The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85: 1135–1148. - PubMed
-
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, et al. (1996) HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381: 667–673. - PubMed
-
- Feng Y, Broder CC, Kennedy PE, Berger EA (1996) HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272: 872–877. - PubMed
-
- Berger EA, Doms RW, Fenyo EM, Korber BT, Littman DR, et al. (1998) A new classification for HIV-1. Nature 391: 240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

