A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria
- PMID: 25313861
- PMCID: PMC4196738
- DOI: 10.1371/journal.pbio.1001969
A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria
Abstract
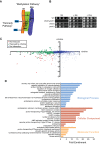
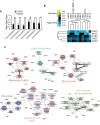
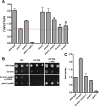
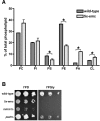
Mitochondrial membrane biogenesis and lipid metabolism require phospholipid transfer from the endoplasmic reticulum (ER) to mitochondria. Transfer is thought to occur at regions of close contact of these organelles and to be nonvesicular, but the mechanism is not known. Here we used a novel genetic screen in S. cerevisiae to identify mutants with defects in lipid exchange between the ER and mitochondria. We show that a strain missing multiple components of the conserved ER membrane protein complex (EMC) has decreased phosphatidylserine (PS) transfer from the ER to mitochondria. Mitochondria from this strain have significantly reduced levels of PS and its derivative phosphatidylethanolamine (PE). Cells lacking EMC proteins and the ER-mitochondria tethering complex called ERMES (the ER-mitochondria encounter structure) are inviable, suggesting that the EMC also functions as a tether. These defects are corrected by expression of an engineered ER-mitochondrial tethering protein that artificially tethers the ER to mitochondria. EMC mutants have a significant reduction in the amount of ER tethered to mitochondria even though ERMES remained intact in these mutants, suggesting that the EMC performs an additional tethering function to ERMES. We find that all Emc proteins interact with the mitochondrial translocase of the outer membrane (TOM) complex protein Tom5 and this interaction is important for PS transfer and cell growth, suggesting that the EMC forms a tether by associating with the TOM complex. Together, our findings support that the EMC tethers ER to mitochondria, which is required for phospholipid synthesis and cell growth.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER.J Lipid Res. 2017 Mar;58(3):553-562. doi: 10.1194/jlr.M072959. Epub 2017 Jan 24. J Lipid Res. 2017. PMID: 28119445 Free PMC article.
-
Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance.Traffic. 2012 Jun;13(6):880-90. doi: 10.1111/j.1600-0854.2012.01352.x. Epub 2012 Apr 8. Traffic. 2012. PMID: 22409400 Free PMC article.
-
ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae.J Cell Sci. 2012 Oct 15;125(Pt 20):4791-9. doi: 10.1242/jcs.105635. Epub 2012 Jul 13. J Cell Sci. 2012. PMID: 22797914 Free PMC article.
-
Connection of Protein Transport and Organelle Contact Sites in Mitochondria.J Mol Biol. 2017 Jul 7;429(14):2148-2160. doi: 10.1016/j.jmb.2017.05.023. Epub 2017 May 30. J Mol Biol. 2017. PMID: 28576471 Review.
-
The ERMES complex and ER-mitochondria connections.Biochem Soc Trans. 2012 Apr;40(2):445-50. doi: 10.1042/BST20110758. Biochem Soc Trans. 2012. PMID: 22435828 Review.
Cited by
-
Interpretable Machine Learning Reveals Dissimilarities Between Subtypes of Autism Spectrum Disorder.Front Genet. 2021 Feb 25;12:618277. doi: 10.3389/fgene.2021.618277. eCollection 2021. Front Genet. 2021. PMID: 33719335 Free PMC article.
-
The phosphatidylethanolamine-binding protein DTH1 mediates degradation of lipid droplets in Chlamydomonas reinhardtii.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):23131-23139. doi: 10.1073/pnas.2005600117. Epub 2020 Aug 31. Proc Natl Acad Sci U S A. 2020. PMID: 32868427 Free PMC article.
-
Lipin1 deficiency causes sarcoplasmic reticulum stress and chaperone-responsive myopathy.EMBO J. 2019 Jan 3;38(1):e99576. doi: 10.15252/embj.201899576. Epub 2018 Nov 12. EMBO J. 2019. PMID: 30420558 Free PMC article.
-
Squaring the EMC - how promoting membrane protein biogenesis impacts cellular functions and organismal homeostasis.J Cell Sci. 2020 Apr 24;133(8):jcs243519. doi: 10.1242/jcs.243519. J Cell Sci. 2020. PMID: 32332093 Free PMC article. Review.
-
A protein quality control pathway at the mitochondrial outer membrane.Elife. 2020 Mar 2;9:e51065. doi: 10.7554/eLife.51065. Elife. 2020. PMID: 32118579 Free PMC article.
References
-
- Elbaz Y, Schuldiner M (2011) Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci 36: 616–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

