Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice
- PMID: 25315009
- PMCID: PMC4375077
- DOI: 10.2337/db14-1238
Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice
Abstract
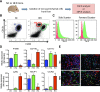
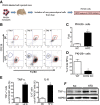
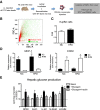
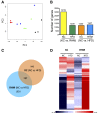
The current dogma is that obesity-associated hepatic inflammation is due to increased Kupffer cell (KC) activation. However, recruited hepatic macrophages (RHMs) were recently shown to represent a sizable liver macrophage population in the context of obesity. Therefore, we assessed whether KCs and RHMs, or both, represent the major liver inflammatory cell type in obesity. We used a combination of in vivo macrophage tracking methodologies and adoptive transfer techniques in which KCs and RHMs are differentially labeled with fluorescent markers. With these approaches, the inflammatory phenotype of these distinct macrophage populations was determined under lean and obese conditions. In vivo macrophage tracking revealed an approximately sixfold higher number of RHMs in obese mice than in lean mice, whereas the number of KCs was comparable. In addition, RHMs comprised smaller size and immature, monocyte-derived cells compared with KCs. Furthermore, RHMs from obese mice were more inflamed and expressed higher levels of tumor necrosis factor-α and interleukin-6 than RHMs from lean mice. A comparison of the MCP-1/C-C chemokine receptor type 2 (CCR2) chemokine system between the two cell types showed that the ligand (MCP-1) is more highly expressed in KCs than in RHMs, whereas CCR2 expression is approximately fivefold greater in RHMs. We conclude that KCs can participate in obesity-induced inflammation by causing the recruitment of RHMs, which are distinct from KCs and are not precursors to KCs. These RHMs then enhance the severity of obesity-induced inflammation and hepatic insulin resistance.
© 2015 by the American Diabetes Association. Readers may use this article as long as the work is properly cited, the use is educational and not for profit, and the work is not altered.
Figures
References
-
- Olefsky JM, Courtney CH. Type 2 diabetes mellitus: etiology, pathogenesis, and natural history. In Endocrinology. 5th ed. DeGroot LJ, Jameson JL, Eds. Philadelphia, W.B. Saunders and Company, 2005, p. 1093–1117
-
- Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860–867 - PubMed
-
- Curat CA, Wegner V, Sengenès C, et al. . Macrophages in human visceral adipose tissue: increased accumulation in obesity and a source of resistin and visfatin. Diabetologia 2006;49:744–747 - PubMed
-
- Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 2004;25:4–7 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL088093/HL/NHLBI NIH HHS/United States
- P30-NS-047101/NS/NINDS NIH HHS/United States
- R24 DK090962/DK/NIDDK NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- DK-033651/DK/NIDDK NIH HHS/United States
- P30-DK-063491/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- W 1226/FWF_/Austrian Science Fund FWF/Austria
- DK-074868/DK/NIDDK NIH HHS/United States
- DK-063491/DK/NIDDK NIH HHS/United States
- R37 DK033651/DK/NIDDK NIH HHS/United States
- F32 DK009062/DK/NIDDK NIH HHS/United States
- P01 DK074868/DK/NIDDK NIH HHS/United States
- R01 DK033651/DK/NIDDK NIH HHS/United States
- DK-09062/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

