Epigenetic disruptions of histone signatures for the trophectoderm and inner cell mass in mouse parthenogenetic embryos
- PMID: 25315067
- PMCID: PMC4333321
- DOI: 10.1089/scd.2014.0310
Epigenetic disruptions of histone signatures for the trophectoderm and inner cell mass in mouse parthenogenetic embryos
Abstract
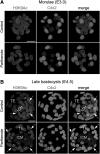
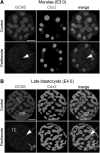
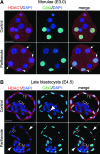
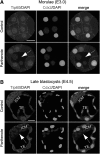
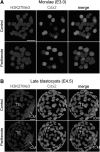
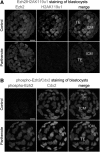
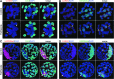
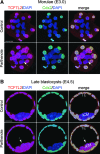
Epigenetic asymmetry has been shown to be associated with the first lineage allocation event in preimplantation development, that is, the formation of the trophectoderm (TE) and inner cell mass (ICM) lineages in the blastocyst. Since parthenogenesis causes aberrant segregation between the TE and ICM lineages, we examined several development-associated histone modifications in parthenotes, including those involved in (i) transcriptional activation [acetylated histone H3 lysine 9 (H3K9Ac) and lysine 14 (H3K14Ac), trimethylated histone H3 lysine 4 (H3K4Me3), and dimethylated histone H3 arginine 26 (H3R26Me2)] and (ii) transcriptional repression [trimethylated histone H3 lysine 9 (H3K9Me3) and lysine 27 (H3K27Me3), and mono-ubiquitinated histone H2A lysine 119 (H2AK119u1)]. Here, we report that in parthenotes, H3R26Me2 expression decreased from the morula stage, while expression patterns and levels of H3K9Ac, H3K27Me3, and H2AK119u1 were unchanged until the blastocyst stage; whereas H3K14Ac, H3K4Me3, and H3K9Me3 showed normal patterns and levels of expressions. Relative to the decrease of H3K9Ac in the ICM and increase in the TE of parthenotes, we detected reduced expression of TAT-interactive protein 60 acetyltransferase and histone deacetylase 1 deacetylase in the ICM and TE of parthenotes, respectively. Relative to the decrease of H3R26Me2, we also observed decreased expression of coactivator-associated arginine methyltransferase 1 methyltransferase and increased expression of the Wnt effector transcription factor 7L2 and miR-181c microRNA in parthenotes. Furthermore, relative to the decrease in H3K27Me3 and H2AK119u1, we found increased phosphorylation of Akt1 and enhancer of zeste homolog 2 in parthenogenetic TE. Therefore, our findings that histone signatures are impaired in parthenotes provide a mechanistic explanation for aberrant lineage segregation and TE defects.
Figures


Similar articles
-
Epigenetic impairments in development of parthenogenetic preimplantation mouse embryos.J Reprod Dev. 2019 Feb 8;65(1):83-90. doi: 10.1262/jrd.2018-028. Epub 2018 Dec 29. J Reprod Dev. 2019. PMID: 30606958 Free PMC article.
-
Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation.J Biol Chem. 2016 Feb 26;291(9):4523-36. doi: 10.1074/jbc.M115.677930. Epub 2016 Jan 5. J Biol Chem. 2016. PMID: 26733201 Free PMC article.
-
Resveratrol modulates epigenetic regulators of promoter histone methylation and acetylation that restores BRCA1, p53, p21CIP1 in human breast cancer cell lines.Biofactors. 2019 Sep;45(5):818-829. doi: 10.1002/biof.1544. Epub 2019 Jul 17. Biofactors. 2019. PMID: 31317586
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
-
The role of histone modifications and variants in regulating gene expression in breast cancer.J Mammary Gland Biol Neoplasia. 2010 Mar;15(1):19-33. doi: 10.1007/s10911-010-9167-z. Epub 2010 Feb 4. J Mammary Gland Biol Neoplasia. 2010. PMID: 20131086 Review.
Cited by
-
The Dynamics of Histone Modifications during Mammalian Zygotic Genome Activation.Int J Mol Sci. 2024 Jan 25;25(3):1459. doi: 10.3390/ijms25031459. Int J Mol Sci. 2024. PMID: 38338738 Free PMC article. Review.
-
Long non-coding RNAs potentially function synergistically in the cellular reprogramming of SCNT embryos.BMC Genomics. 2018 Aug 23;19(1):631. doi: 10.1186/s12864-018-5021-2. BMC Genomics. 2018. PMID: 30139326 Free PMC article.
-
Epigenetic impairments in development of parthenogenetic preimplantation mouse embryos.J Reprod Dev. 2019 Feb 8;65(1):83-90. doi: 10.1262/jrd.2018-028. Epub 2018 Dec 29. J Reprod Dev. 2019. PMID: 30606958 Free PMC article.
References
-
- Cassar G, John TM. and Etches RJ. (1998). Observations on ploidy of cells and on reproductive performance in parthenogenetic turkeys. Poult Sci 77:1457–1462 - PubMed
-
- Fujita MK. and Moritz C. (2009). Origin and evolution of parthenogenetic genomes in lizards: current state and future directions. Cytogenet Genome Res 127:261–272 - PubMed
-
- Parker HM, Kiess AS, Wells JB, Young KM, Rowe D. and McDaniel CD. (2010). Genetic selection increases parthenogenesis in Chinese painted quail (Coturnix chinensis). Poult Sci 89:1468–1472 - PubMed
-
- Sinclair EA, Pramuk JB, Bezy RL, Crandall KA. and Sites JW, Jr., (2010). DNA evidence for nonhybrid origins of parthenogenesis in natural populations of vertebrates. Evolution 64:1346–1357 - PubMed
-
- Cheng L. (2008). More new lines of human parthenogenetic embryonic stem cells. Cell Res 18:215–217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

