The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-not) complex to promote elongation
- PMID: 25315781
- PMCID: PMC4246073
- DOI: 10.1074/jbc.C114.601088
The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-not) complex to promote elongation
Abstract
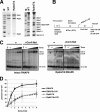
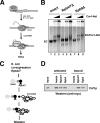
Gene expression relies on the balance between mRNA synthesis in the nucleus and decay in the cytoplasm, processes once thought to be separate. We now know that transcription and decay rates are coordinated, but the factors or molecular mechanisms are unclear. The Ccr4-Not complex regulates multiple stages of gene expression, from mRNA synthesis to protein destruction. One of its functions is to promote RNA polymerase II elongation by reactivating arrested elongation complexes. Here we explored the features of polymerase required for Ccr4-Not to promote elongation and found that the Rpb4/7 module is important for Ccr4-Not to associate with elongation complexes and stimulate elongation. Rpb4/7 has also been implicated in coordinating mRNA synthesis and decay, but its role in this process is controversial. The interplay between Ccr4-Not and Rpb4/7 described here suggests a mechanism for how the cell coordinates mRNA synthesis and decay.
Keywords: Ccr4-Not; RNA Polymerase II; RNA Synthesis; Transcription Elongation Factor; Transcription Regulation; mRNA Decay.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Pérez-Ortín J. E., de Miguel-Jiménez L., Chávez S. (2012) Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim. Biophys. Acta 1819, 604–615 - PubMed
-
- Sun M., Schwalb B., Pirkl N., Maier K. C., Schenk A., Failmezger H., Tresch A., Cramer P. (2013) Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 52, 52–62 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

