GEN1 from a thermophilic fungus is functionally closely similar to non-eukaryotic junction-resolving enzymes
- PMID: 25315822
- PMCID: PMC4270448
- DOI: 10.1016/j.jmb.2014.10.008
GEN1 from a thermophilic fungus is functionally closely similar to non-eukaryotic junction-resolving enzymes
Abstract
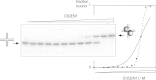
Processing of Holliday junctions is essential in recombination. We have identified the gene for the junction-resolving enzyme GEN1 from the thermophilic fungus Chaetomium thermophilum and expressed the N-terminal 487-amino-acid section. The protein is a nuclease that is highly selective for four-way DNA junctions, cleaving 1nt 3' to the point of strand exchange on two strands symmetrically disposed about a diagonal axis. CtGEN1 binds to DNA junctions as a discrete homodimer with nanomolar affinity. Analysis of the kinetics of cruciform cleavage shows that cleavage of the second strand occurs an order of magnitude faster than the first cleavage so as to generate a productive resolution event. All these properties are closely similar to those described for bacterial, phage and mitochondrial junction-resolving enzymes. CtGEN1 is also similar in properties to the human enzyme but lacks the problems with aggregation that currently prevent detailed analysis of the latter protein. CtGEN1 is thus an excellent enzyme with which to engage in biophysical and structural analysis of eukaryotic GEN1.
Keywords: Chaetomium thermophilum; DNA recombination and repair; FEN1; Holliday junction resolution; thermophilic proteins.
Copyright © 2014. Published by Elsevier Ltd.
Figures






Similar articles
-
A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.Nucleic Acids Res. 2018 Nov 16;46(20):11089-11098. doi: 10.1093/nar/gky863. Nucleic Acids Res. 2018. PMID: 30247722 Free PMC article.
-
Biochemical and Structural Properties of Fungal Holliday Junction-Resolving Enzymes.Methods Enzymol. 2018;600:543-568. doi: 10.1016/bs.mie.2017.11.021. Epub 2018 Feb 1. Methods Enzymol. 2018. PMID: 29458774
-
Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.Cell Rep. 2015 Dec 22;13(11):2565-2575. doi: 10.1016/j.celrep.2015.11.042. Epub 2015 Dec 10. Cell Rep. 2015. PMID: 26686639 Free PMC article.
-
Holliday junction-resolving enzymes-structures and mechanisms.FEBS Lett. 2017 Apr;591(8):1073-1082. doi: 10.1002/1873-3468.12529. Epub 2017 Jan 1. FEBS Lett. 2017. PMID: 27990631 Review.
-
The interaction of four-way DNA junctions with resolving enzymes.Biochem Soc Trans. 2010 Apr;38(2):399-403. doi: 10.1042/BST0380399. Biochem Soc Trans. 2010. PMID: 20298191 Review.
Cited by
-
Canonical and novel non-canonical activities of the Holliday junction resolvase Yen1.Nucleic Acids Res. 2022 Jan 11;50(1):259-280. doi: 10.1093/nar/gkab1225. Nucleic Acids Res. 2022. PMID: 34928393 Free PMC article.
-
Substrate preference of Gen endonucleases highlights the importance of branched structures as DNA damage repair intermediates.Nucleic Acids Res. 2017 May 19;45(9):5333-5348. doi: 10.1093/nar/gkx214. Nucleic Acids Res. 2017. PMID: 28369583 Free PMC article.
-
MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.Nat Commun. 2024 Jun 17;15(1):5140. doi: 10.1038/s41467-024-49490-9. Nat Commun. 2024. PMID: 38886375 Free PMC article.
-
A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.Nucleic Acids Res. 2018 Nov 16;46(20):11089-11098. doi: 10.1093/nar/gky863. Nucleic Acids Res. 2018. PMID: 30247722 Free PMC article.
-
Search and processing of Holliday junctions within long DNA by junction-resolving enzymes.Nat Commun. 2022 Oct 7;13(1):5921. doi: 10.1038/s41467-022-33503-6. Nat Commun. 2022. PMID: 36207294 Free PMC article.
References
-
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. - PubMed
-
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. - PubMed
-
- Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. - PubMed
-
- Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am J Dis Child. 1954;88:754–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

