Long-time evolution and highly dynamic satellite DNA in leptodactylid and hylodid frogs
- PMID: 25316286
- PMCID: PMC4201667
- DOI: 10.1186/s12863-014-0111-x
Long-time evolution and highly dynamic satellite DNA in leptodactylid and hylodid frogs
Abstract
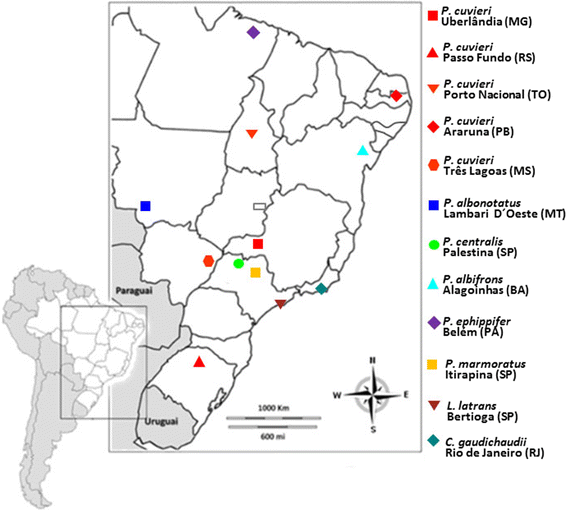
Background: Satellite DNA sequences are the most abundant components of heterochromatin and are repeated in tandem hundreds to thousands of times in the genome. However, the number of repeats of a specific satellite family can vary even between the genomes of related species or populations. The PcP190 satellite DNA family was identified in the genome of the leptodactylid frog Physalaemus cuvieri, which showed to be derived most likely from the 5S rDNA in an ancestral species. In this study, we investigate the presence of the PcP190 satellite DNA in several P. cuvieri populations and in four closely related species at the chromosomal and molecular level. Furthermore, we investigate the occurrence of this satellite DNA in the genomes of P. marmoratus as well as in representative species of the leptodactylid genus Leptodactylus (L. latrans) and the hylodid family (Crossodactylus gaudichaudii), all with the aim of investigating if the PcP190 satellite DNA presents or not a restricted distribution.
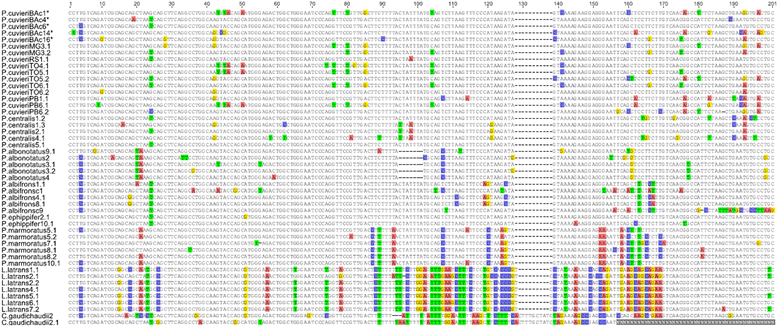
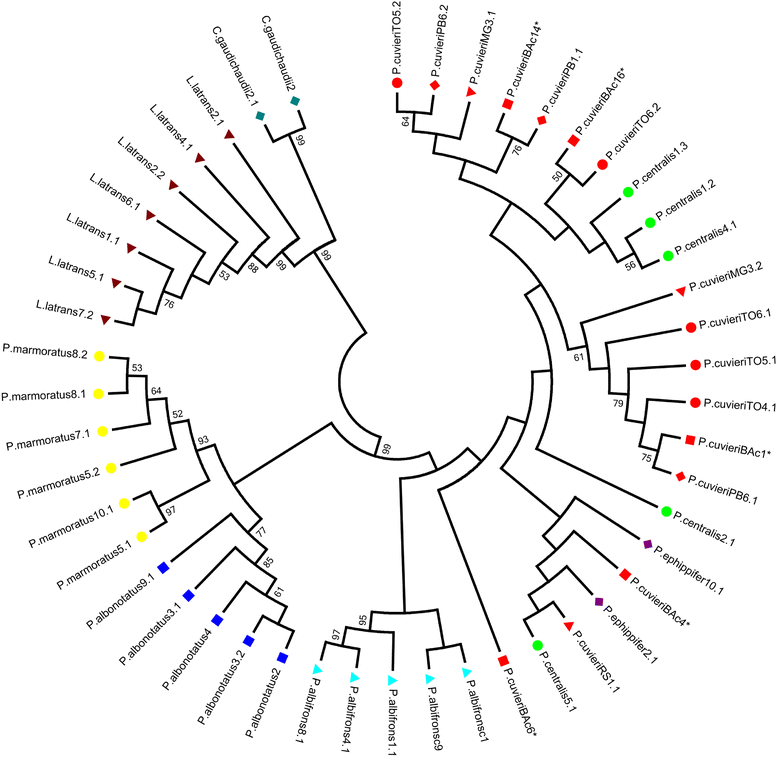
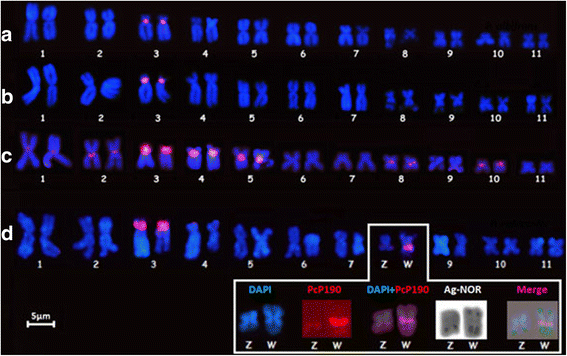
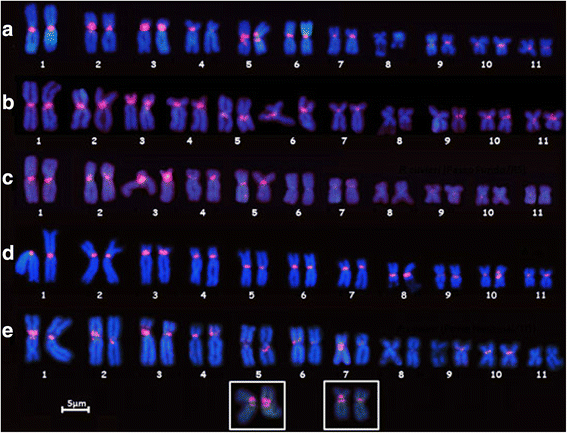
Results: The PcP190 satellite DNA was detected in all the analyzed species. Some of them exhibited particular sequence differences, allowing the identification of species-specific groups of sequences, but in other species, the sequences were more conserved. However, in a general analysis, conserved and variable domains have been recognized within the PcP190 monomer. The chromosomal analysis performed on P. cuvieri populations and closely related species revealed high variability of the satellite DNA amount and its chromosomal location, which has always been coincident with regions of centromeric/pericentromeric heterochromatin.
Conclusion: The PcP190 satellite DNA was found in representatives of two families, Leptodactylidae and Hylodidae, indicating that these sequences are widely distributed and conserved in these frogs. There is a pattern of non-random variation within the repeating units, indicating interplay between stochastic events and selective pressure along the PcP190 sequences. Karyotypic differences involving the PcP190 satellite DNA prove to be highly dynamic on the chromosomes of the Physalaemus and its differential accumulation has contributed to the differentiation process of the Z and W sex chromosomes in P. ephippifer.
Figures
Similar articles
-
Cytogenetic characterization and mapping of the repetitive DNAs in Cycloramphus bolitoglossus (Werner, 1897): More clues for the chromosome evolution in the genus Cycloramphus (Anura, Cycloramphidae).PLoS One. 2021 Jan 13;16(1):e0245128. doi: 10.1371/journal.pone.0245128. eCollection 2021. PLoS One. 2021. PMID: 33439901 Free PMC article.
-
High diversity of 5S ribosomal DNA and evidence of recombination with the satellite DNA PcP190 in frogs.Gene. 2023 Jan 30;851:147015. doi: 10.1016/j.gene.2022.147015. Epub 2022 Oct 29. Gene. 2023. PMID: 36374718
-
New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content.Biomolecules. 2025 Jun 16;15(6):876. doi: 10.3390/biom15060876. Biomolecules. 2025. PMID: 40563516 Free PMC article.
-
Satellite DNA and chromosomes in Neotropical fishes: methods, applications and perspectives.J Fish Biol. 2010 Apr;76(5):1094-116. doi: 10.1111/j.1095-8649.2010.02564.x. J Fish Biol. 2010. PMID: 20409164 Review.
-
Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin.Gene. 2008 Feb 15;409(1-2):72-82. doi: 10.1016/j.gene.2007.11.013. Epub 2007 Dec 4. Gene. 2008. PMID: 18182173 Review.
Cited by
-
Intra-generic and interspecific karyotype patterns of Leptodactylus and Adenomera (Anura, Leptodactylidae) with inclusion of five species from Central Amazonia.Genetica. 2016 Feb;144(1):37-46. doi: 10.1007/s10709-015-9876-8. Epub 2015 Dec 9. Genetica. 2016. PMID: 26650375
-
Evidence for the Transcription of a Satellite DNA Widely Found in Frogs.Genes (Basel). 2024 Dec 5;15(12):1572. doi: 10.3390/genes15121572. Genes (Basel). 2024. PMID: 39766839 Free PMC article.
-
Satellite DNA Mapping in Pseudis fusca (Hylidae, Pseudinae) Provides New Insights into Sex Chromosome Evolution in Paradoxical Frogs.Genes (Basel). 2019 Feb 19;10(2):160. doi: 10.3390/genes10020160. Genes (Basel). 2019. PMID: 30791490 Free PMC article.
-
Cytogenetic and genetic data support Crossodactylus aeneus Müller, 1924 as a new junior synonym of C. gaudichaudii Duméril and Bibron, 1841 (Amphibia, Anura).Genet Mol Biol. 2021 Mar 22;44(2):e20200301. doi: 10.1590/1678-4685-GMB-2020-0301. eCollection 2021. Genet Mol Biol. 2021. PMID: 33751017 Free PMC article.
-
Transcription of tandemly repetitive DNA: functional roles.Chromosome Res. 2015 Sep;23(3):463-77. doi: 10.1007/s10577-015-9494-4. Chromosome Res. 2015. PMID: 26403245 Review.
References
-
- López-Flores I, Ramos-Garrido MA. The repetitive DNA content of eukaryotic genomes. In: López-Flores I, editor. Repetitive DNA. Granada: Karger; 2012. pp. 1–28. - PubMed
-
- Plohl M, Meštrović N, Mravinac B. Satellite DNA evolution. In: López-Flores I, editor. Repetitive DNA. Granada: Karger; 2012. pp. 126–152. - PubMed
-
- Tsoumani KT, Drosopoulou E, Mavragani-Tsipidou P, Mathiopoulos KD. Molecular characterization and chromosomal distribution of a species-specific transcribed centromeric satellite repeat from the olive fruit Fly, Bactrocera oleae. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0079393. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

