Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City
- PMID: 25316698
- PMCID: PMC4205793
- DOI: 10.1128/mBio.01933-14
Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City
Abstract
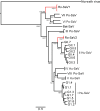
Norway rats (Rattus norvegicus) are globally distributed and concentrate in urban environments, where they live and feed in closer proximity to human populations than most other mammals. Despite the potential role of rats as reservoirs of zoonotic diseases, the microbial diversity present in urban rat populations remains unexplored. In this study, we used targeted molecular assays to detect known bacterial, viral, and protozoan human pathogens and unbiased high-throughput sequencing to identify novel viruses related to agents of human disease in commensal Norway rats in New York City. We found that these rats are infected with bacterial pathogens known to cause acute or mild gastroenteritis in people, including atypical enteropathogenic Escherichia coli, Clostridium difficile, and Salmonella enterica, as well as infectious agents that have been associated with undifferentiated febrile illnesses, including Bartonella spp., Streptobacillus moniliformis, Leptospira interrogans, and Seoul hantavirus. We also identified a wide range of known and novel viruses from groups that contain important human pathogens, including sapoviruses, cardioviruses, kobuviruses, parechoviruses, rotaviruses, and hepaciviruses. The two novel hepaciviruses discovered in this study replicate in the liver of Norway rats and may have utility in establishing a small animal model of human hepatitis C virus infection. The results of this study demonstrate the diversity of microbes carried by commensal rodent species and highlight the need for improved pathogen surveillance and disease monitoring in urban environments. Importance: The observation that most emerging infectious diseases of humans originate in animal reservoirs has led to wide-scale microbial surveillance and discovery programs in wildlife, particularly in the developing world. Strikingly, less attention has been focused on commensal animals like rats, despite their abundance in urban centers and close proximity to human populations. To begin to explore the zoonotic disease risk posed by urban rat populations, we trapped and surveyed Norway rats collected in New York City over a 1-year period. This analysis revealed a striking diversity of known pathogens and novel viruses in our study population, including multiple agents associated with acute gastroenteritis or febrile illnesses in people. Our findings indicate that urban rats are reservoirs for a vast diversity of microbes that may affect human health and indicate a need for increased surveillance and awareness of the disease risks associated with urban rodent infestation.
Copyright © 2014 Firth et al.
Figures



References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

