Nanoparticle encapsidation of Flock house virus by auto assembly of Tobacco mosaic virus coat protein
- PMID: 25318056
- PMCID: PMC4227231
- DOI: 10.3390/ijms151018540
Nanoparticle encapsidation of Flock house virus by auto assembly of Tobacco mosaic virus coat protein
Abstract
Tobacco Mosaic virus (TMV) coat protein is well known for its ability to self-assemble into supramolecular nanoparticles, either as protein discs or as rods originating from the ~300 bp genomic RNA origin-of-assembly (OA). We have utilized TMV self-assembly characteristics to create a novel Flock House virus (FHV) RNA nanoparticle. FHV encodes a viral polymerase supporting autonomous replication of the FHV genome, which makes it an attractive candidate for viral transgene expression studies and targeted RNA delivery into host cells. However, FHV viral genome size is strictly limited by native FHV capsid. To determine if this packaging restriction could be eliminated, FHV was adapted to express enhanced green fluorescent protein (GFP), to allow for monitoring of functional FHV RNA activity. Then TMV OA was introduced in six 3' insertion sites, with only site one supporting functional FHV GFP expression. To create nanoparticles, FHV GFP-OA modified genomic RNA was mixed in vitro with TMV coat protein and monitored for encapsidation by agarose electrophoresis and electron microscopy. The production of TMV-like rod shaped nanoparticles indicated that modified FHV RNA can be encapsidated by purified TMV coat protein by self-assembly. This is the first demonstration of replication-independent packaging of the FHV genome by protein self-assembly.
Figures
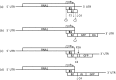
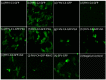


Similar articles
-
In planta production of flock house virus transencapsidated RNA and its potential use as a vaccine.Mol Biotechnol. 2015 Apr;57(4):325-36. doi: 10.1007/s12033-014-9826-1. Mol Biotechnol. 2015. PMID: 25432792
-
Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors.Virology. 2003 Jan 5;305(1):10-24. doi: 10.1006/viro.2002.1740. Virology. 2003. PMID: 12504536
-
Plant Expression of Trans-Encapsidated Chimeric Viral Vaccines with Animal RNA Replicons: An Update.Methods Mol Biol. 2024;2786:289-300. doi: 10.1007/978-1-0716-3770-8_13. Methods Mol Biol. 2024. PMID: 38814400
-
Plant Expression of Trans-Encapsidated Viral Nanoparticle Vaccines with Animal RNA Replicons.Methods Mol Biol. 2017;1499:77-86. doi: 10.1007/978-1-4939-6481-9_4. Methods Mol Biol. 2017. PMID: 27987143 Review.
-
Recent insights into the biology and biomedical applications of Flock House virus.Cell Mol Life Sci. 2008 Sep;65(17):2675-87. doi: 10.1007/s00018-008-8037-y. Cell Mol Life Sci. 2008. PMID: 18516498 Free PMC article. Review.
Cited by
-
The pharmacology of plant virus nanoparticles.Virology. 2021 Apr;556:39-61. doi: 10.1016/j.virol.2021.01.012. Epub 2021 Jan 28. Virology. 2021. PMID: 33545555 Free PMC article. Review.
-
Engineered Flock House Virus for Targeted Gene Suppression Through RNAi in Fruit Flies (Drosophila melanogaster) in Vitro and in Vivo.Front Physiol. 2018 Jul 3;9:805. doi: 10.3389/fphys.2018.00805. eCollection 2018. Front Physiol. 2018. PMID: 30018564 Free PMC article.
-
Plant Virus Expression Vectors: A Powerhouse for Global Health.Biomedicines. 2017 Jul 30;5(3):44. doi: 10.3390/biomedicines5030044. Biomedicines. 2017. PMID: 28758953 Free PMC article. Review.
-
A Self-Amplifying Human Papillomavirus 16 Vaccine Candidate Delivered by Tobacco Mosaic Virus-Like Particles.ACS Appl Bio Mater. 2024 Nov 18;7(11):7675-7683. doi: 10.1021/acsabm.4c01239. Epub 2024 Nov 8. ACS Appl Bio Mater. 2024. PMID: 39512153 Free PMC article.
-
Polymer-mediated gene therapy: Recent advances and merging of delivery techniques.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020 Mar;12(2):e1598. doi: 10.1002/wnan.1598. Epub 2019 Dec 2. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020. PMID: 31793237 Free PMC article. Review.
References
-
- Turner D.R., Joyce L.E., Butler P.J.G. The Tobacco Mosaic virus assembly origin RNA: Functional characteristics defined by directed mutagenesis. J. Mol. Biol. 1988;203:531–547. - PubMed
-
- Gallie D.R., Sleat D.E., Watts J.W., Turner P.C., Wilson T.M.A. In vivo uncoating and efficient expression of foreign mrnas packaged in tmv-like particles. Science. 1987;236:1122–1124. - PubMed
-
- Smith M.L., Corbo T., Bernales J., Lindbo J.A., Pogue G.P., Palmer K.E., McCormick A.A. Assembly of trans-encapsidated recombinant viral vectors engineered from Tobacco Mosaic virus and semliki forest virus and their evaluation as immunogens. Virology. 2007;358:321–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

