Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season
- PMID: 25318541
- PMCID: PMC4287553
- DOI: 10.1186/1471-2164-15-899
Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season
Abstract
Background: Seasonal estrus is a critical limiting factor of animal fecundity, and it involves changes in both ovarian biology and hormone secretion in different seasons. Previous studies indicate that two classes of small RNAs (miRNAs and piRNAs) play important regulatory roles in ovarian biology. To understand the roles of small RNA-mediated post-transcriptional regulation in ovine seasonal estrus, the variation in expression patterns of ovarian small RNAs during anestrus and the breeding season were analyzed using Solexa sequencing technology. In addition, reproductive hormone levels were determined during ovine anestrus and the breeding season.
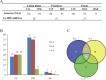
Results: A total of 483 miRNAs (including 97 known, 369 conserved and 17 predicated novel miRNAs), which belong to 183 different miRNA families, were identified in ovaries of Tan sheep and Small Tail Han (STH) sheep. Compared with the three stages of the breeding season, 25 shared significantly differentially expressed (including 19 up- and six down-regulated) miRNAs were identified in ovine anestrus. KEGG Pathway analysis revealed that the target genes for some of the differentially expressed miRNAs were involved in reproductive hormone related pathways (e.g. steroid biosynthesis, androgen and estrogen metabolism and GnRH signaling pathway) as well as follicular/luteal development related pathways. Moreover, the expression of the differentially expressed miRNAs and most of their target genes were negatively correlated in the above pathways. Furthermore, the levels of estrogen, progesterone and LH in ovine anestrus were significantly lower than those in the breeding season. Combining the results of pathway enrichment analysis, expression of target genes and hormone measurement, we suggest that these differentially expressed miRNAs in anestrus might participate in attenuation of ovarian activity by regulating the above pathways. Besides miRNAs, a large and unexpectedly diverse set of piRNAs were also identified.
Conclusions: The miRNA profiles of ovine ovaries in anestrus were presented for the first time. The identification and characterization of miRNAs that are differentially expressed between ovine anestrus and the breeding season will help understanding of the role of miRNAs in the regulation of seasonal estrus, and provides candidates for determining miRNAs which could be potentially used to regulate ovine seasonal estrus.
Figures






Similar articles
-
Comparative profiling of differentially expressed microRNAs in estrous ovaries of Kazakh sheep in different seasons.Gene. 2018 Jul 20;664:181-191. doi: 10.1016/j.gene.2018.04.025. Epub 2018 Apr 25. Gene. 2018. PMID: 29704632
-
Expression characteristics of pineal miRNAs at ovine different reproductive stages and the identification of miRNAs targeting the AANAT gene.BMC Genomics. 2021 Mar 25;22(1):217. doi: 10.1186/s12864-021-07536-y. BMC Genomics. 2021. PMID: 33765915 Free PMC article.
-
Transcriptome Analysis of Neuroendocrine Regulation of Ovine Hypothalamus-Pituitary-Ovary Axis during Ovine Anestrus and the Breeding Season.Genes (Basel). 2021 Nov 24;12(12):1861. doi: 10.3390/genes12121861. Genes (Basel). 2021. PMID: 34946810 Free PMC article.
-
[The molecular mechanism of sheep seasonal breeding and artificial regulatory techniques for estrus and mating in anestrus].Yi Chuan. 2018 May 20;40(5):369-377. doi: 10.16288/j.yczz.17-423. Yi Chuan. 2018. PMID: 29785945 Review. Chinese.
-
The postpartum buffalo. II. Acyclicity and anestrus.Anim Reprod Sci. 2007 Feb;97(3-4):216-36. doi: 10.1016/j.anireprosci.2006.03.003. Epub 2006 Apr 18. Anim Reprod Sci. 2007. PMID: 16621354 Review.
Cited by
-
MicroRNA Mediating Networks in Granulosa Cells Associated with Ovarian Follicular Development.Biomed Res Int. 2017;2017:4585213. doi: 10.1155/2017/4585213. Epub 2017 Feb 19. Biomed Res Int. 2017. PMID: 28316977 Free PMC article.
-
Non-coding RNA regulation in reproduction: Their potential use as biomarkers.Noncoding RNA Res. 2019 May 3;4(2):54-62. doi: 10.1016/j.ncrna.2019.04.001. eCollection 2019 Jun. Noncoding RNA Res. 2019. PMID: 31193491 Free PMC article. Review.
-
MicroRNA 221 expression in theca and granulosa cells: hormonal regulation and function.J Anim Sci. 2018 Mar 6;96(2):641-652. doi: 10.1093/jas/skx069. J Anim Sci. 2018. PMID: 29385487 Free PMC article.
-
Comparative mRNA and miRNA expression in European mouflon (Ovis musimon) and sheep (Ovis aries) provides novel insights into the genetic mechanisms for female reproductive success.Heredity (Edinb). 2019 Feb;122(2):172-186. doi: 10.1038/s41437-018-0090-1. Epub 2018 May 21. Heredity (Edinb). 2019. PMID: 29784930 Free PMC article.
-
Insight Into Pituitary lncRNA and mRNA at Two Estrous Stages in Small Tail Han Sheep With Different FecB Genotypes.Front Endocrinol (Lausanne). 2022 Feb 1;12:789564. doi: 10.3389/fendo.2021.789564. eCollection 2021. Front Endocrinol (Lausanne). 2022. PMID: 35178025 Free PMC article.
References
-
- Tu YR. Sheep and Goat Breeds in China: Shanghai Scientific & Technology Publishers. 1989.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

