Epigenomics of macrophages
- PMID: 25319330
- PMCID: PMC4203424
- DOI: 10.1111/imr.12213
Epigenomics of macrophages
Abstract
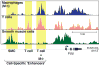
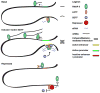
Macrophages play essential roles in tissue homeostasis, pathogen elimination, and tissue repair. A defining characteristic of these cells is their ability to efficiently adapt to a variety of abruptly changing and complex environments. This ability is intrinsically linked to a capacity to quickly alter their transcriptome, and this is tightly associated with the epigenomic organization of these cells and, in particular, their enhancer repertoire. Indeed, enhancers are genomic sites that serve as platforms for the integration of signaling pathways with the mechanisms that regulate mRNA transcription. Notably, transcription is pervasive at active enhancers and enhancer RNAs (eRNAs) are tightly coupled to regulated transcription of protein-coding genes. Furthermore, given that each cell type possesses a defining enhancer repertoire, studies on enhancers provide a powerful method to study how specialization of functions among the diverse macrophage subtypes may arise. Here, we review recent studies providing insights into the distinct mechanisms that contribute to the establishment of enhancers and their role in the regulation of transcription in macrophages.
Keywords: PU.1; eRNA; enhancer; epigenetic; macrophage; transcription.
© 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures

References
-
- Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

