Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function
- PMID: 25319829
- PMCID: PMC4201289
- DOI: 10.1101/gad.242313.114
Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function
Abstract
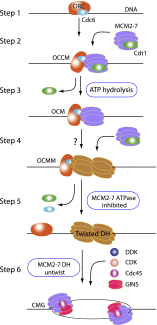
Eukaryotic cells license each DNA replication origin during G1 phase by assembling a prereplication complex that contains a Mcm2-7 (minichromosome maintenance proteins 2-7) double hexamer. During S phase, each Mcm2-7 hexamer forms the core of a replicative DNA helicase. However, the mechanisms of origin licensing and helicase activation are poorly understood. The helicase loaders ORC-Cdc6 function to recruit a single Cdt1-Mcm2-7 heptamer to replication origins prior to Cdt1 release and ORC-Cdc6-Mcm2-7 complex formation, but how the second Mcm2-7 hexamer is recruited to promote double-hexamer formation is not well understood. Here, structural evidence for intermediates consisting of an ORC-Cdc6-Mcm2-7 complex and an ORC-Cdc6-Mcm2-7-Mcm2-7 complex are reported, which together provide new insights into DNA licensing. Detailed structural analysis of the loaded Mcm2-7 double-hexamer complex demonstrates that the two hexamers are interlocked and misaligned along the DNA axis and lack ATP hydrolysis activity that is essential for DNA helicase activity. Moreover, we show that the head-to-head juxtaposition of the Mcm2-7 double hexamer generates a new protein interaction surface that creates a multisubunit-binding site for an S-phase protein kinase that is known to activate DNA replication. The data suggest how the double hexamer is assembled and how helicase activity is regulated during DNA licensing, with implications for cell cycle control of DNA replication and genome stability.
Keywords: DNA replication initiation; electron microscopy; origin recognition complex; prereplication complex; replicative helicase.
© 2014 Sun et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Arias EE, Walter JC. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 - PubMed
-
- Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 - PubMed
-
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 - PubMed
-
- Bochman ML, Schwacha A. 2008. The Mcm2–7 complex has in vitro helicase activity. Mol Cell 31: 287–293 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
