Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis
- PMID: 25319994
- PMCID: PMC4248312
- DOI: 10.1101/gr.164830.113
Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis
Abstract
We used mouse ENCODE data along with complementary data from other laboratories to study the dynamics of occupancy and the role in gene regulation of the transcription factor TAL1, a critical regulator of hematopoiesis, at multiple stages of hematopoietic differentiation. We combined ChIP-seq and RNA-seq data in six mouse cell types representing a progression from multilineage precursors to differentiated erythroblasts and megakaryocytes. We found that sites of occupancy shift dramatically during commitment to the erythroid lineage, vary further during terminal maturation, and are strongly associated with changes in gene expression. In multilineage progenitors, the likely target genes are enriched for hematopoietic growth and functions associated with the mature cells of specific daughter lineages (such as megakaryocytes). In contrast, target genes in erythroblasts are specifically enriched for red cell functions. Furthermore, shifts in TAL1 occupancy during erythroid differentiation are associated with gene repression (dissociation) and induction (co-occupancy with GATA1). Based on both enrichment for transcription factor binding site motifs and co-occupancy determined by ChIP-seq, recruitment by GATA transcription factors appears to be a stronger determinant of TAL1 binding to chromatin than the canonical E-box binding site motif. Studies of additional proteins lead to the model that TAL1 regulates expression after being directed to a distinct subset of genomic binding sites in each cell type via its association with different complexes containing master regulators such as GATA2, ERG, and RUNX1 in multilineage cells and the lineage-specific master regulator GATA1 in erythroblasts.
© 2014 Wu et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

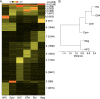

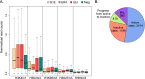




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- U01 HL099656/HL/NHLBI NIH HHS/United States
- P30 DK090969/DK/NIDDK NIH HHS/United States
- R56 DK065806/DK/NIDDK NIH HHS/United States
- R37DK058044/DK/NIDDK NIH HHS/United States
- U01HL099656/HL/NHLBI NIH HHS/United States
- P30DK090969/DK/NIDDK NIH HHS/United States
- R37 DK058044/DK/NIDDK NIH HHS/United States
- U01 HL099993/HL/NHLBI NIH HHS/United States
- R01DK065806/DK/NIDDK NIH HHS/United States
- R01DK58044/DK/NIDDK NIH HHS/United States
- U54 HG006998/HG/NHGRI NIH HHS/United States
- R01DK54937/DK/NIDDK NIH HHS/United States
- RC2 HG005573/HG/NHGRI NIH HHS/United States
- R01 DK058044/DK/NIDDK NIH HHS/United States
- R01 DK065806/DK/NIDDK NIH HHS/United States
- RC2HG005573/HG/NHGRI NIH HHS/United States
- U54HG006998/HG/NHGRI NIH HHS/United States
- R01 DK054937/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
