ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors
- PMID: 25320010
- PMCID: PMC4300142
- DOI: 10.1158/0008-5472.CAN-14-2073
ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors
Abstract
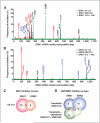
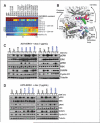
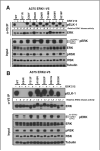
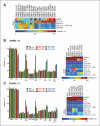
The use of targeted therapeutics directed against BRAF(V600)-mutant metastatic melanoma improves progression-free survival in many patients; however, acquired drug resistance remains a major medical challenge. By far, the most common clinical resistance mechanism involves reactivation of the MAPK (RAF/MEK/ERK) pathway by a variety of mechanisms. Thus, targeting ERK itself has emerged as an attractive therapeutic concept, and several ERK inhibitors have entered clinical trials. We sought to preemptively determine mutations in ERK1/2 that confer resistance to either ERK inhibitors or combined RAF/MEK inhibition in BRAF(V600)-mutant melanoma. Using a random mutagenesis screen, we identified multiple point mutations in ERK1 (MAPK3) and ERK2 (MAPK1) that could confer resistance to ERK or RAF/MEK inhibitors. ERK inhibitor-resistant alleles were sensitive to RAF/MEK inhibitors and vice versa, suggesting that the future development of alternating RAF/MEK and ERK inhibitor regimens might help circumvent resistance to these agents.
©2014 American Association for Cancer Research.
Figures



Similar articles
-
Protein Kinase CK2α Maintains Extracellular Signal-regulated Kinase (ERK) Activity in a CK2α Kinase-independent Manner to Promote Resistance to Inhibitors of RAF and MEK but Not ERK in BRAF Mutant Melanoma.J Biol Chem. 2016 Aug 19;291(34):17804-15. doi: 10.1074/jbc.M115.712885. Epub 2016 May 17. J Biol Chem. 2016. PMID: 27226552 Free PMC article.
-
PAK signalling drives acquired drug resistance to MAPK inhibitors in BRAF-mutant melanomas.Nature. 2017 Oct 5;550(7674):133-136. doi: 10.1038/nature24040. Epub 2017 Sep 27. Nature. 2017. PMID: 28953887 Free PMC article.
-
RAF-Mutant Melanomas Differentially Depend on ERK2 Over ERK1 to Support Aberrant MAPK Pathway Activation and Cell Proliferation.Mol Cancer Res. 2021 Jun;19(6):1063-1075. doi: 10.1158/1541-7786.MCR-20-1022. Epub 2021 Mar 11. Mol Cancer Res. 2021. PMID: 33707308
-
MAP kinase signaling and inhibition in melanoma.Oncogene. 2013 May 9;32(19):2373-9. doi: 10.1038/onc.2012.345. Epub 2012 Sep 3. Oncogene. 2013. PMID: 22945644 Review.
-
Clinical Development of BRAF plus MEK Inhibitor Combinations.Trends Cancer. 2020 Sep;6(9):797-810. doi: 10.1016/j.trecan.2020.05.009. Epub 2020 Jun 13. Trends Cancer. 2020. PMID: 32540454 Review.
Cited by
-
A novel chemical genetic approach reveals paralog-specific role of ERK1/2 in mouse embryonic stem cell fate control.Front Cell Dev Biol. 2024 Jul 12;12:1415621. doi: 10.3389/fcell.2024.1415621. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39071800 Free PMC article.
-
Spatial and temporal alterations in protein structure by EGF regulate cryptic cysteine oxidation.Sci Signal. 2020 Jan 21;13(615):eaay7315. doi: 10.1126/scisignal.aay7315. Sci Signal. 2020. PMID: 31964804 Free PMC article.
-
Preclinical Anticipation of On- and Off-Target Resistance Mechanisms to Anti-Cancer Drugs: A Systematic Review.Int J Mol Sci. 2024 Jan 5;25(2):705. doi: 10.3390/ijms25020705. Int J Mol Sci. 2024. PMID: 38255778 Free PMC article.
-
Oncogenic KRAS blockade therapy: renewed enthusiasm and persistent challenges.Mol Cancer. 2021 Oct 4;20(1):128. doi: 10.1186/s12943-021-01422-7. Mol Cancer. 2021. PMID: 34607583 Free PMC article. Review.
-
Erk1R84H is an oncoprotein that causes hepatocellular carcinoma in mice and imposes a rigorous negative feedback loop.Oncogene. 2025 Aug;44(31):2689-2714. doi: 10.1038/s41388-025-03437-6. Epub 2025 May 20. Oncogene. 2025. PMID: 40394416 Free PMC article.
References
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. - PubMed
-
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

