The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia
- PMID: 25320243
- PMCID: PMC4347284
- DOI: 10.1182/blood-2014-05-577270
The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia
Abstract
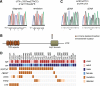
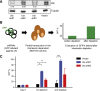
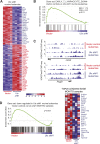
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive form of leukemia that is mainly diagnosed in children and shows a skewed gender distribution toward males. In this study, we report somatic loss-of-function mutations in the X-linked histone H3K27me3 demethylase ubiquitously transcribed X (UTX) chromosome, in human T-ALL. Interestingly, UTX mutations were exclusively present in male T-ALL patients and allelic expression analysis revealed that UTX escapes X-inactivation in female T-ALL lymphoblasts and normal T cells. Notably, we demonstrate in vitro and in vivo that the H3K27me3 demethylase UTX functions as a bona fide tumor suppressor in T-ALL. Moreover, T-ALL driven by UTX inactivation exhibits collateral sensitivity to pharmacologic H3K27me3 inhibition. All together, our results show how a gender-specific and therapeutically relevant defect in balancing H3K27 methylation contributes to T-cell leukemogenesis.
© 2015 by The American Society of Hematology.
Figures


Comment in
-
An X-linked tumor suppressor in T-ALL.Blood. 2015 Jan 1;125(1):3-4. doi: 10.1182/blood-2014-11-607838. Blood. 2015. PMID: 25554741 No abstract available.
References
-
- Goldberg JM, Silverman LB, Levy DE, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21(19):3616–3622. - PubMed
-
- Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. - PubMed
-
- Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19(4):484–497. - PubMed
-
- Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 2005;106(1):274–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

