The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection
- PMID: 25320327
- PMCID: PMC4301124
- DOI: 10.1128/JVI.01913-14
The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection
Abstract
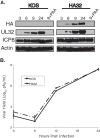
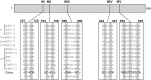
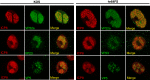
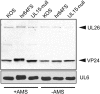
During DNA encapsidation, herpes simplex virus 1 (HSV-1) procapsids are converted to DNA-containing capsids by a process involving activation of the viral protease, expulsion of the scaffold proteins, and the uptake of viral DNA. Encapsidation requires six minor capsid proteins (UL6, UL15, UL17, UL25, UL28, and UL33) and one viral protein, UL32, not found to be associated with capsids. Although functions have been assigned to each of the minor capsid proteins, the role of UL32 in encapsidation has remained a mystery. Using an HSV-1 variant containing a functional hemagglutinin-tagged UL32, we demonstrated that UL32 was synthesized with true late kinetics and that it exhibited a previously unrecognized localization pattern. At 6 to 9 h postinfection (hpi), UL32 accumulated in viral replication compartments in the nucleus of the host cell, while at 24 hpi, it was additionally found in the cytoplasm. A newly generated UL32-null mutant was used to confirm that although B capsids containing wild-type levels of capsid proteins were synthesized, these procapsids were unable to initiate the encapsidation process. Furthermore, we showed that UL32 is redox sensitive and identified two highly conserved oxidoreductase-like C-X-X-C motifs that are essential for protein function. In addition, the disulfide bond profiles of the viral proteins UL6, UL25, and VP19C and the viral protease, VP24, were altered in the absence of UL32, suggesting that UL32 may act to modulate disulfide bond formation during procapsid assembly and maturation.
Importance: Although functions have been assigned to six of the seven required packaging proteins of HSV, the role of UL32 in encapsidation has remained a mystery. UL32 is a cysteine-rich viral protein that contains C-X-X-C motifs reminiscent of those in proteins that participate in the regulation of disulfide bond formation. We have previously demonstrated that disulfide bonds are required for the formation and stability of the viral capsids and are also important for the formation and stability of the UL6 portal ring. In this report, we demonstrate that the disulfide bond profiles of the viral proteins UL6, UL25, and VP19C and the viral protease, VP24, are altered in cells infected with a newly isolated UL32-null mutant virus, suggesting that UL32 acts as a chaperone capable of modulating disulfide bond formation. Furthermore, these results suggest that proper regulation of disulfide bonds is essential for initiating encapsidation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging.J Virol. 1998 Sep;72(9):7428-39. doi: 10.1128/JVI.72.9.7428-7439.1998. J Virol. 1998. PMID: 9696839 Free PMC article.
-
Disulfide bond formation in the herpes simplex virus 1 UL6 protein is required for portal ring formation and genome encapsidation.J Virol. 2011 Sep;85(17):8616-24. doi: 10.1128/JVI.00123-11. Epub 2011 May 18. J Virol. 2011. PMID: 21593161 Free PMC article.
-
The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged.Virology. 1998 Dec 5;252(1):115-25. doi: 10.1006/viro.1998.9439. Virology. 1998. PMID: 9875322
-
Herpesvirus Capsid Assembly and DNA Packaging.Adv Anat Embryol Cell Biol. 2017;223:119-142. doi: 10.1007/978-3-319-53168-7_6. Adv Anat Embryol Cell Biol. 2017. PMID: 28528442 Free PMC article. Review.
-
The Terminase Complex of Each Human Herpesvirus.Biol Pharm Bull. 2024;47(5):912-916. doi: 10.1248/bpb.b23-00717. Biol Pharm Bull. 2024. PMID: 38692868 Review.
Cited by
-
Structures and Divergent Mechanisms in Capsid Maturation and Stabilization Following Genome Packaging of Human Cytomegalovirus and Herpesviruses.Life (Basel). 2021 Feb 16;11(2):150. doi: 10.3390/life11020150. Life (Basel). 2021. PMID: 33669389 Free PMC article. Review.
-
Large Subunit of the Human Herpes Simplex Virus Terminase as a Promising Target in Design of Anti-Herpesvirus Agents.Molecules. 2023 Oct 31;28(21):7375. doi: 10.3390/molecules28217375. Molecules. 2023. PMID: 37959793 Free PMC article.
-
The Exonuclease Activity of Herpes Simplex Virus 1 UL12 Is Required for Production of Viral DNA That Can Be Packaged To Produce Infectious Virus.J Virol. 2017 Nov 14;91(23):e01380-17. doi: 10.1128/JVI.01380-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28956767 Free PMC article.
-
The Proteome and Secretome of Cortical Brain Cells Infected With Herpes Simplex Virus.Front Neurol. 2020 Aug 27;11:844. doi: 10.3389/fneur.2020.00844. eCollection 2020. Front Neurol. 2020. PMID: 32973653 Free PMC article.
-
Epstein-Barr virus genome packaging factors accumulate in BMRF1-cores within viral replication compartments.PLoS One. 2019 Sep 13;14(9):e0222519. doi: 10.1371/journal.pone.0222519. eCollection 2019. PLoS One. 2019. PMID: 31518362 Free PMC article.
References
-
- Brown J, McVoy M, Homa F. 2002. Packaging DNA into herpesvirus capsids, p 111–153. In Holzenburg A, Bogner E (ed), Structure-function relationships of human pathogenic viruses. Springer, New York, NY.
-
- Baines J, Weller S. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p 135–150. In Viral genome packaging machines: genetics, structure, and mechanism. Springer, New York, NY.
-
- Conway JF, Homa FL. 2011. Nucleocapsid structure, assembly and DNA packaging of herpes simplex virus. Caister Academic Press, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

