The ubiquitination machinery of the ubiquitin system
- PMID: 25320573
- PMCID: PMC4196676
- DOI: 10.1199/tab.0174
The ubiquitination machinery of the ubiquitin system
Abstract
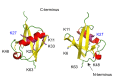
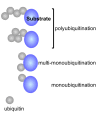
The protein ubiquitin is a covalent modifier of proteins, including itself. The ubiquitin system encompasses the enzymes required for catalysing attachment of ubiquitin to substrates as well as proteins that bind to ubiquitinated proteins leading them to their final fate. Also included are activities that remove ubiquitin independent of, or in concert with, proteolysis of the substrate, either by the proteasome or proteases in the vacuole. In addition to ubiquitin encoded by a family of fusion proteins, there are proteins with ubiquitin-like domains, likely forming ubiquitin's β-grasp fold, but incapable of covalent modification. However, they serve as protein-protein interaction platforms within the ubiquitin system. Multi-gene families encode all of these types of activities. Within the ubiquitination machinery "half" of the ubiquitin system are redundant, partially redundant, and unique components affecting diverse developmental and environmental responses in plants. Notably, multiple aspects of biotic and abiotic stress responses require, or are modulated by, ubiquitination. Finally, aspects of the ubiquitin system have broad utility: as components to enhance gene expression or to regulate protein abundance. This review focuses on the ubiquitination machinery: ubiquitin, unique aspects about the synthesis of ubiquitin and organization of its gene family, ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2) and ubiquitin ligases, or E3s. Given the large number of E3s in Arabidopsis this review covers the U box, HECT and RING type E3s, with the exception of the cullin-based E3s.
Figures



References
-
- Acconcia F., Sigismund S., Polo S. Ubiquitin in trafficking: The network at work. Exp. Cell Res. 2009;315:1610–1618. - PubMed
-
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 2004;279:40053–40061. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
