Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes
- PMID: 25323695
- PMCID: PMC4458378
- DOI: 10.1126/science.1249830
Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes
Abstract
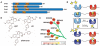
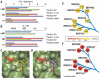
Small molecules are useful tools for probing the biological function and therapeutic potential of individual proteins, but achieving selectivity is challenging when the target protein shares structural domains with other proteins. The Bromo and Extra-Terminal (BET) proteins have attracted interest because of their roles in transcriptional regulation, epigenetics, and cancer. The BET bromodomains (protein interaction modules that bind acetyl-lysine) have been targeted by potent small-molecule inhibitors, but these inhibitors lack selectivity for individual family members. We developed an ethyl derivative of an existing small-molecule inhibitor, I-BET/JQ1, and showed that it binds leucine/alanine mutant bromodomains with nanomolar affinity and achieves up to 540-fold selectivity relative to wild-type bromodomains. Cell culture studies showed that blockade of the first bromodomain alone is sufficient to displace a specific BET protein, Brd4, from chromatin. Expansion of this approach could help identify the individual roles of single BET proteins in human physiology and disease.
Copyright © 2014, American Association for the Advancement of Science.
Figures

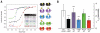
Comment in
-
A bumpy, holey method to probe proteins.Nat Methods. 2015 Jan;12(1):14. doi: 10.1038/nmeth.3246. Nat Methods. 2015. PMID: 25699317 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- 097945/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- BB/G023123/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 100476/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/J001201/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

