P2CS: updates of the prokaryotic two-component systems database
- PMID: 25324303
- PMCID: PMC4384028
- DOI: 10.1093/nar/gku968
P2CS: updates of the prokaryotic two-component systems database
Abstract
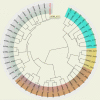
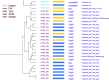
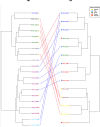
The P2CS database (http://www.p2cs.org/) is a comprehensive resource for the analysis of Prokaryotic Two-Component Systems (TCSs). TCSs are comprised of a receptor histidine kinase (HK) and a partner response regulator (RR) and control important prokaryotic behaviors. The latest incarnation of P2CS includes 164,651 TCS proteins, from 2758 sequenced prokaryotic genomes. Several important new features have been added to P2CS since it was last described. Users can search P2CS via BLAST, adding hits to their cart, and homologous proteins can be aligned using MUSCLE and viewed using Jalview within P2CS. P2CS also provides phylogenetic trees based on the conserved signaling domains of the RRs and HKs from entire genomes. HK and RR trees are annotated with gene organization and domain architecture, providing insights into the evolutionary origin of the contemporary gene set. The majority of TCSs are encoded by adjacent HK and RR genes, however, 'orphan' unpaired TCS genes are also abundant and identifying their partner proteins is challenging. P2CS now provides paired HK and RR trees with proteins from the same genetic locus indicated. This allows the appraisal of evolutionary relationships across entire TCSs and in some cases the identification of candidate partners for orphan TCS proteins.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Whitworth D.E. Two-component regulatory systems in prokaryotes. In: Filloux A, editor. Bacterial Regulatory Networks. Caister: Horizon Press; 2012. pp. 191–222.
-
- Whitworth D.E. Classification and organization of two-component systems. In: Gross R, Beier D, editors. Two-Component Systems. Norfolk: Horizon Scientific Press; 2012. pp. 1–20.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

