PLAZA 3.0: an access point for plant comparative genomics
- PMID: 25324309
- PMCID: PMC4384038
- DOI: 10.1093/nar/gku986
PLAZA 3.0: an access point for plant comparative genomics
Abstract
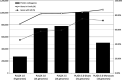
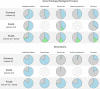
Comparative sequence analysis has significantly altered our view on the complexity of genome organization and gene functions in different kingdoms. PLAZA 3.0 is designed to make comparative genomics data for plants available through a user-friendly web interface. Structural and functional annotation, gene families, protein domains, phylogenetic trees and detailed information about genome organization can easily be queried and visualized. Compared with the first version released in 2009, which featured nine organisms, the number of integrated genomes is more than four times higher, and now covers 37 plant species. The new species provide a wider phylogenetic range as well as a more in-depth sampling of specific clades, and genomes of additional crop species are present. The functional annotation has been expanded and now comprises data from Gene Ontology, MapMan, UniProtKB/Swiss-Prot, PlnTFDB and PlantTFDB. Furthermore, we improved the algorithms to transfer functional annotation from well-characterized plant genomes to other species. The additional data and new features make PLAZA 3.0 (http://bioinformatics.psb.ugent.be/plaza/) a versatile and comprehensible resource for users wanting to explore genome information to study different aspects of plant biology, both in model and non-model organisms.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Van de Peer Y., Pires J.C. Getting up to speed. Curr. Opin. Plant Biol. 2012;15:111–114. - PubMed
-
- Leeggangers H.A., Moreno-Pachon N., Gude H., Immink R.G. Transfer of knowledge about flowering and vegetative propagation from model species to bulbous plants. Int. J. Dev. Biol. 2013;57:611–620. - PubMed
-
- Tian C.F., Zhou Y.J., Zhang Y.M., Li Q.Q., Zhang Y.Z., Li D.F., Wang S., Wang J., Gilbert L.B., Li Y.R., et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl Acad. Sci. U.S.A. 2012;109:8629–8634. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

