Survey of chimeric IStron elements in bacterial genomes: multiple molecular symbioses between group I intron ribozymes and DNA transposons
- PMID: 25324310
- PMCID: PMC4227781
- DOI: 10.1093/nar/gku939
Survey of chimeric IStron elements in bacterial genomes: multiple molecular symbioses between group I intron ribozymes and DNA transposons
Abstract
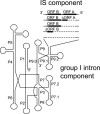
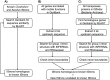
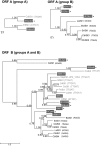
IStrons are chimeric genetic elements composed of a group I intron associated with an insertion sequence (IS). The group I intron is a catalytic RNA providing the IStron with self-splicing ability, which renders IStron insertions harmless to the host genome. The IS element is a DNA transposon conferring mobility, and thus allowing the IStron to spread in genomes. IStrons are therefore a striking example of a molecular symbiosis between unrelated genetic elements endowed with different functions. In this study, we have conducted the first comprehensive survey of IStrons in sequenced genomes that provides insights into the distribution, diversity, origin and evolution of IStrons. We show that IStrons have a restricted phylogenetic distribution limited to two bacterial phyla, the Firmicutes and the Fusobacteria. Nevertheless, diverse IStrons representing two major groups targeting different insertion site motifs were identified. This taken with the finding that while the intron components of all IStrons belong to the same structural class, they are fused to different IS families, indicates that multiple intron-IS symbioses have occurred during evolution. In addition, introns and IS elements related to those that were at the origin of IStrons were also identified.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Frost L.S., Leplae R., Summers A.O., Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. 2005;3:722–732. - PubMed
-
- Hazen T.H., Pan L., Gu J.D., Sobecky P.A. The contribution of mobile genetic elements to the evolution and ecology of Vibrios. FEMS Microbiol. Ecol. 2010;74:485–499. - PubMed
-
- Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Lisch D. How important are transposons for plant evolution. Nat. Rev. Genet. 2013;14:49–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

