Aging and energetics' 'Top 40' future research opportunities 2010-2013
- PMID: 25324965
- PMCID: PMC4197746
- DOI: 10.12688/f1000research.5212.1
Aging and energetics' 'Top 40' future research opportunities 2010-2013
Abstract
Background: As part of a coordinated effort to expand our research activity at the interface of Aging and Energetics a team of investigators at The University of Alabama at Birmingham systematically assayed and catalogued the top research priorities identified in leading publications in that domain, believing the result would be useful to the scientific community at large.
Objective: To identify research priorities and opportunities in the domain of aging and energetics as advocated in the 40 most cited papers related to aging and energetics in the last 4 years.
Design: The investigators conducted a search for papers on aging and energetics in Scopus, ranked the resulting papers by number of times they were cited, and selected the ten most-cited papers in each of the four years that include 2010 to 2013, inclusive.
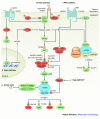
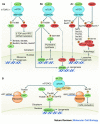
Results: Ten research categories were identified from the 40 papers. These included: (1) Calorie restriction (CR) longevity response, (2) role of mTOR (mechanistic target of Rapamycin) and related factors in lifespan extension, (3) nutrient effects beyond energy (especially resveratrol, omega-3 fatty acids, and selected amino acids), 4) autophagy and increased longevity and health, (5) aging-associated predictors of chronic disease, (6) use and effects of mesenchymal stem cells (MSCs), (7) telomeres relative to aging and energetics, (8) accretion and effects of body fat, (9) the aging heart, and (10) mitochondria, reactive oxygen species, and cellular energetics.
Conclusion: The field is rich with exciting opportunities to build upon our existing knowledge about the relations among aspects of aging and aspects of energetics and to better understand the mechanisms which connect them.
Conflict of interest statement
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

