Hidden treasures in "ancient" microarrays: gene-expression portrays biology and potential resistance pathways of major lung cancer subtypes and normal tissue
- PMID: 25325012
- PMCID: PMC4178426
- DOI: 10.3389/fonc.2014.00251
Hidden treasures in "ancient" microarrays: gene-expression portrays biology and potential resistance pathways of major lung cancer subtypes and normal tissue
Abstract
Objective: Novel statistical methods and increasingly more accurate gene annotations can transform "old" biological data into a renewed source of knowledge with potential clinical relevance. Here, we provide an in silico proof-of-concept by extracting novel information from a high-quality mRNA expression dataset, originally published in 2001, using state-of-the-art bioinformatics approaches.
Methods: The dataset consists of histologically defined cases of lung adenocarcinoma (AD), squamous (SQ) cell carcinoma, small-cell lung cancer, carcinoid, metastasis (breast and colon AD), and normal lung specimens (203 samples in total). A battery of statistical tests was used for identifying differential gene expressions, diagnostic and prognostic genes, enriched gene ontologies, and signaling pathways.
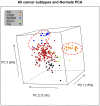
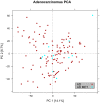
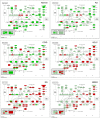
Results: Our results showed that gene expressions faithfully recapitulate immunohistochemical subtype markers, as chromogranin A in carcinoids, cytokeratin 5, p63 in SQ, and TTF1 in non-squamous types. Moreover, biological information with putative clinical relevance was revealed as potentially novel diagnostic genes for each subtype with specificity 93-100% (AUC = 0.93-1.00). Cancer subtypes were characterized by (a) differential expression of treatment target genes as TYMS, HER2, and HER3 and (b) overrepresentation of treatment-related pathways like cell cycle, DNA repair, and ERBB pathways. The vascular smooth muscle contraction, leukocyte trans-endothelial migration, and actin cytoskeleton pathways were overexpressed in normal tissue.
Conclusion: Reanalysis of this public dataset displayed the known biological features of lung cancer subtypes and revealed novel pathways of potentially clinical importance. The findings also support our hypothesis that even old omics data of high quality can be a source of significant biological information when appropriate bioinformatics methods are used.
Keywords: bioinformatics; carcinoid; lung adenocarcinoma; mesothelioma; microarray; small cell; squamous.
Figures





References
-
- Smit EF, Socinski MA, Mullaney BP, Myrand SP, Scagliotti GV, Lorigan P, et al. Biomarker analysis in a phase III study of pemetrexed-carboplatin versus etoposide-carboplatin in chemonaive patients with extensive-stage small-cell lung cancer. Ann Oncol (2012) 23:1723–910.1093/annonc/mdr563 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

