A PDB-wide, evolution-based assessment of protein-protein interfaces
- PMID: 25326082
- PMCID: PMC4274722
- DOI: 10.1186/s12900-014-0022-0
A PDB-wide, evolution-based assessment of protein-protein interfaces
Abstract
Background: Thanks to the growth in sequence and structure databases, more than 50 million sequences are now available in UniProt and 100,000 structures in the PDB. Rich information about protein-protein interfaces can be obtained by a comprehensive study of protein contacts in the PDB, their sequence conservation and geometric features.
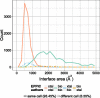
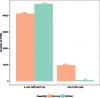
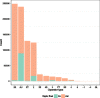
Results: An automated computational pipeline was developed to run our Evolutionary Protein-Protein Interface Classifier (EPPIC) software on the entire PDB and store the results in a relational database, currently containing > 800,000 interfaces. This allows the analysis of interface data on a PDB-wide scale. Two large benchmark datasets of biological interfaces and crystal contacts, each containing about 3000 entries, were automatically generated based on criteria thought to be strong indicators of interface type. The BioMany set of biological interfaces includes NMR dimers solved as crystal structures and interfaces that are preserved across diverse crystal forms, as catalogued by the Protein Common Interface Database (ProtCID) from Xu and Dunbrack. The second dataset, XtalMany, is derived from interfaces that would lead to infinite assemblies and are therefore crystal contacts. BioMany and XtalMany were used to benchmark the EPPIC approach. The performance of EPPIC was also compared to classifications from the Protein Interfaces, Surfaces, and Assemblies (PISA) program on a PDB-wide scale, finding that the two approaches give the same call in about 88% of PDB interfaces. By comparing our safest predictions to the PDB author annotations, we provide a lower-bound estimate of the error rate of biological unit annotations in the PDB. Additionally, we developed a PyMOL plugin for direct download and easy visualization of EPPIC interfaces for any PDB entry. Both the datasets and the PyMOL plugin are available at http://www.eppic-web.org/ewui/\#downloads.
Conclusions: Our computational pipeline allows us to analyze protein-protein contacts and their sequence conservation across the entire PDB. Two new benchmark datasets are provided, which are over an order of magnitude larger than existing manually curated ones. These tools enable the comprehensive study of several aspects of protein-protein contacts in the PDB and represent a basis for future, even larger scale studies of protein-protein interactions.
Figures


References
-
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242,. doi: 10.1093/nar/28.1.235. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=102472&too... - DOI - PMC - PubMed
-
- http://doi.wiley.com/10.1002/prot.22787 Schärer Ma, Grütter MG, Capitani G: CRK: An evolutionary approach for distinguishing biologically relevant interfaces from crystal contacts. Proteins: Struct Funct Bioinformatics2010,[] - DOI - PubMed
-
- Duarte JM, Srebniak A, Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinformatics. 2012;13:334,. doi: 10.1186/1471-2105-13-334. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3556496&to... - DOI - PMC - PubMed
-
- Duarte JM, Biyani N, Baskaran K, Capitani G. An analysis of oligomerization interfaces in transmembrane proteins. BMC Struct Biol. 2013;13:21. doi: 10.1186/1472-6807-13-21. [http://www.ncbi.nlm.nih.gov/pubmed/24134166] - DOI - PMC - PubMed
-
- Ivan G, Szabadka Z, Grolmusz V. A hybrid clustering of protein binding sites. Febs J. 2010;277(6):1494–1502. doi: 10.1111/j.1742-4658.2010.07578.x. [http://www.ncbi.nlm.nih.gov/pubmed/20148971] - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

