Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426
- PMID: 25326311
- PMCID: PMC4272721
- DOI: 10.1128/AEM.02577-14
Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426
Abstract
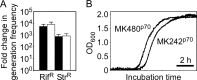
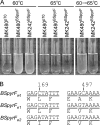
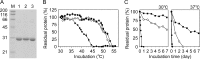
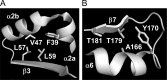
Thermostability is an important property of enzymes utilized for practical applications because it allows long-term storage and use as catalysts. In this study, we constructed an error-prone strain of the thermophile Geobacillus kaustophilus HTA426 and investigated thermoadaptation-directed enzyme evolution using the strain. A mutation frequency assay using the antibiotics rifampin and streptomycin revealed that G. kaustophilus had substantially higher mutability than Escherichia coli and Bacillus subtilis. The predominant mutations in G. kaustophilus were A · T→G · C and C · G→T · A transitions, implying that the high mutability of G. kaustophilus was attributable in part to high-temperature-associated DNA damage during growth. Among the genes that may be involved in DNA repair in G. kaustophilus, deletions of the mutSL, mutY, ung, and mfd genes markedly enhanced mutability. These genes were subsequently deleted to construct an error-prone thermophile that showed much higher (700- to 9,000-fold) mutability than the parent strain. The error-prone strain was auxotrophic for uracil owing to the fact that the strain was deficient in the intrinsic pyrF gene. Although the strain harboring Bacillus subtilis pyrF was also essentially auxotrophic, cells became prototrophic after 2 days of culture under uracil starvation, generating B. subtilis PyrF variants with an enhanced half-denaturation temperature of >10°C. These data suggest that this error-prone strain is a promising host for thermoadaptation-directed evolution to generate thermostable variants from thermolabile enzymes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
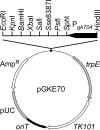
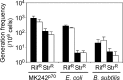
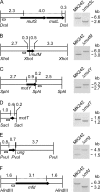
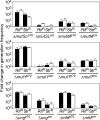
References
-
- Nakamura A, Takakura Y, Kobayashi H, Hoshino T. 2005. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J Biosci Bioeng 100:158–163. doi:10.1263/jbb.100.158. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

