Identification of a Mg2+-sensitive ORF in the 5'-leader of TRPM7 magnesium channel mRNA
- PMID: 25326319
- PMCID: PMC4227784
- DOI: 10.1093/nar/gku951
Identification of a Mg2+-sensitive ORF in the 5'-leader of TRPM7 magnesium channel mRNA
Abstract
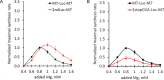
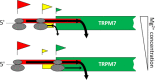
TRPM7 is an essential and ubiquitous channel-kinase regulating cellular influx of Mg2+. Although TRPM7 mRNA is highly abundant, very small amount of the protein is detected in cells, suggesting post-transcriptional regulation of trpm7 gene expression. We found that TRPM7 mRNA 5'-leader contains two evolutionarily conserved upstream open reading frames that act together to drastically inhibit translation of the TRPM7 reading frame at high magnesium levels and ensure its optimal translation at low magnesium levels, when the activity of the channel-kinase is most required. The study provides the first example of magnesium channel synthesis being controlled by Mg2+ in higher eukaryotes.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


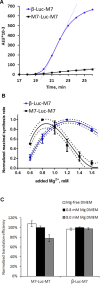
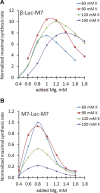
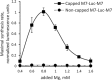
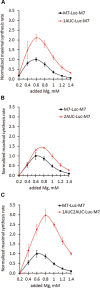
References
-
- Nadler M.J., Hermosura M.C., Inabe K., Perraud A.L., Zhu Q., Stokes A.J., Kurosaki T., Kinet J.P., Penner R., Scharenberg A.M., et al. LTRPC7 is a Mg·ATP-regulated divalent cation channel required for cell viability. Nature. 2001;411:590–595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

