Mechanistic insights into allosteric structure-function relationships at the M1 muscarinic acetylcholine receptor
- PMID: 25326383
- PMCID: PMC4246120
- DOI: 10.1074/jbc.M114.604967
Mechanistic insights into allosteric structure-function relationships at the M1 muscarinic acetylcholine receptor
Abstract
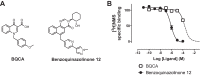
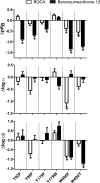
Benzylquinolone carboxylic acid (BQCA) is the first highly selective positive allosteric modulator (PAM) for the M1 muscarinic acetylcholine receptor (mAChR), but it possesses low affinity for the allosteric site on the receptor. More recent drug discovery efforts identified 3-((1S,2S)-2-hydroxycyclohexyl)-6-((6-(1-methyl-1H-pyrazol-4-yl)pyridin-3-yl)methyl)benzo[h]quinazolin-4(3H)-one (referred to herein as benzoquinazolinone 12) as a more potent M1 mAChR PAM with a structural ancestry originating from BQCA and related compounds. In the current study, we optimized the synthesis of and fully characterized the pharmacology of benzoquinazolinone 12, finding that its improved potency derived from a 50-fold increase in allosteric site affinity as compared with BQCA, while retaining a similar level of positive cooperativity with acetylcholine. We then utilized site-directed mutagenesis and molecular modeling to validate the allosteric binding pocket we previously described for BQCA as a shared site for benzoquinazolinone 12 and provide a molecular basis for its improved activity at the M1 mAChR. This includes a key role for hydrophobic and polar interactions with residues Tyr-179, in the second extracellular loop (ECL2) and Trp-400(7.35) in transmembrane domain (TM) 7. Collectively, this study highlights how the properties of affinity and cooperativity can be differentially modified on a common structural scaffold and identifies molecular features that can be exploited to tailor the development of M1 mAChR-targeting PAMs.
Keywords: Allosteric Regulation; Cell Signaling; Drug Discovery; G Protein-coupled Receptor (GPCR); Muscarinic Acetylcholine Receptor; Mutagenesis.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Byun N. E., Grannan M., Bubser M., Barry R. L., Thompson A., Rosanelli J., Gowrishankar R., Kelm N. D., Damon S., Bridges T. M., Melancon B. J., Tarr J. C., Brogan J. T., Avison M. J., Deutch A. Y., Wess J., Wood M. R., Lindsley C. W., Gore J. C., Conn P. J., Jones C. K. (2014) Antipsychotic drug-like effects of the selective M4 muscarinic acetylcholine receptor positive allosteric modulator VU0152100. Neuropsychopharmacology 39, 1578–1593 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

