Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability
- PMID: 25326701
- PMCID: PMC4244265
- DOI: 10.1038/ng.3120
Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability
Abstract
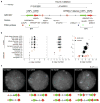
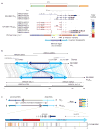
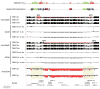
Recurrent deletions of chromosome 15q13.3 associate with intellectual disability, schizophrenia, autism and epilepsy. To gain insight into the instability of this region, we sequenced it in affected individuals, normal individuals and nonhuman primates. We discovered five structural configurations of the human chromosome 15q13.3 region ranging in size from 2 to 3 Mb. These configurations arose recently (∼0.5-0.9 million years ago) as a result of human-specific expansions of segmental duplications and two independent inversion events. All inversion breakpoints map near GOLGA8 core duplicons-a ∼14-kb primate-specific chromosome 15 repeat that became organized into larger palindromic structures. GOLGA8-flanked palindromes also demarcate the breakpoints of recurrent 15q13.3 microdeletions, the expansion of chromosome 15 segmental duplications in the human lineage and independent structural changes in apes. The significant clustering (P = 0.002) of breakpoints provides mechanistic evidence for the role of this core duplicon and its palindromic architecture in promoting the evolutionary and disease-related instability of chromosome 15.
Conflict of interest statement
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and was an SAB member of Pacific Biosciences, Inc. (2009–2013) and SynapDx Corp. (2011–2013). J.A.R. is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer, Inc. L.G.S. was an employee of Signature Genomic Laboratories and is now an employee of Genetic Veterinary Sciences, Inc.
Figures




References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

