Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation
- PMID: 25327251
- PMCID: PMC4968649
- DOI: 10.1038/nature13901
Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation
Abstract
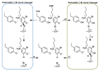
Organohalide chemistry underpins many industrial and agricultural processes, and a large proportion of environmental pollutants are organohalides. Nevertheless, organohalide chemistry is not exclusively of anthropogenic origin, with natural abiotic and biological processes contributing to the global halide cycle. Reductive dehalogenases are responsible for biological dehalogenation in organohalide respiring bacteria, with substrates including polychlorinated biphenyls or dioxins. Reductive dehalogenases form a distinct subfamily of cobalamin (B12)-dependent enzymes that are usually membrane associated and oxygen sensitive, hindering detailed studies. Here we report the characterization of a soluble, oxygen-tolerant reductive dehalogenase and, by combining structure determination with EPR (electron paramagnetic resonance) spectroscopy and simulation, show that a direct interaction between the cobalamin cobalt and the substrate halogen underpins catalysis. In contrast to the carbon-cobalt bond chemistry catalysed by the other cobalamin-dependent subfamilies, we propose that reductive dehalogenases achieve reduction of the organohalide substrate via halogen-cobalt bond formation. This presents a new model in both organohalide and cobalamin (bio)chemistry that will guide future exploitation of these enzymes in bioremediation or biocatalysis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


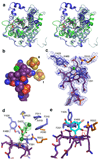


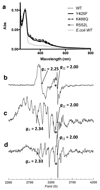

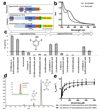
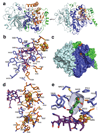

Similar articles
-
Heterologous production and biophysical characterization of catabolic Nitratireductor pacificus pht-3B reductive dehalogenase.Methods Enzymol. 2022;668:327-347. doi: 10.1016/bs.mie.2022.01.004. Epub 2022 Feb 1. Methods Enzymol. 2022. PMID: 35589200
-
A common mechanism for coenzyme cobalamin-dependent reductive dehalogenases.Phys Chem Chem Phys. 2017 Feb 22;19(8):6090-6094. doi: 10.1039/c6cp08659d. Phys Chem Chem Phys. 2017. PMID: 28191552
-
Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.J Biol Chem. 2015 Nov 13;290(46):27572-81. doi: 10.1074/jbc.M115.685693. Epub 2015 Sep 16. J Biol Chem. 2015. PMID: 26378237 Free PMC article.
-
Biochemistry of Catabolic Reductive Dehalogenation.Annu Rev Biochem. 2017 Jun 20;86:357-386. doi: 10.1146/annurev-biochem-061516-044829. Annu Rev Biochem. 2017. PMID: 28654328 Review.
-
Organohalide Respiring Bacteria and Reductive Dehalogenases: Key Tools in Organohalide Bioremediation.Front Microbiol. 2016 Mar 1;7:249. doi: 10.3389/fmicb.2016.00249. eCollection 2016. Front Microbiol. 2016. PMID: 26973626 Free PMC article. Review.
Cited by
-
Transformation of the recalcitrant pesticide chlordecone by Desulfovibrio sp.86 with a switch from ring-opening dechlorination to reductive sulfidation activity.Sci Rep. 2020 Aug 11;10(1):13545. doi: 10.1038/s41598-020-70124-9. Sci Rep. 2020. PMID: 32782344 Free PMC article.
-
Accurate prediction by AlphaFold2 for ligand binding in a reductive dehalogenase and implications for PFAS (per- and polyfluoroalkyl substance) biodegradation.Sci Rep. 2023 Mar 11;13(1):4082. doi: 10.1038/s41598-023-30310-x. Sci Rep. 2023. PMID: 36906658 Free PMC article.
-
Cobalamin Riboswitches Are Broadly Sensitive to Corrinoid Cofactors to Enable an Efficient Gene Regulatory Strategy.mBio. 2022 Oct 26;13(5):e0112122. doi: 10.1128/mbio.01121-22. Epub 2022 Aug 22. mBio. 2022. PMID: 35993747 Free PMC article.
-
Dehalogenases: From Improved Performance to Potential Microbial Dehalogenation Applications.Molecules. 2018 May 7;23(5):1100. doi: 10.3390/molecules23051100. Molecules. 2018. PMID: 29735886 Free PMC article. Review.
-
TsrM as a Model for Purifying and Characterizing Cobalamin-Dependent Radical S-Adenosylmethionine Methylases.Methods Enzymol. 2017;595:303-329. doi: 10.1016/bs.mie.2017.07.007. Epub 2017 Aug 21. Methods Enzymol. 2017. PMID: 28882204 Free PMC article.
References
-
- Stringer Ruth, Johnston Paul., editors. Chlorine and the Environment: An overview of the chlorine industry. Dordrecht, the Netherlands: Kluwer Academic; 2001. p. 448. ISBN 0792367979.
-
- Oberg G. The natural chlorine cycle--fitting the scattered pieces. Appl Microbiol Biotechnol. 2002;58:565–81. - PubMed
-
- Gribble GW. Occurrence of halogenated alkaloids. Alkaloids Chem Biol. 2012;71:1–165. - PubMed
-
- Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 206080/ERC_/European Research Council/International
- BB/C00521X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E013007/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H021523/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

