Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal
- PMID: 25327252
- PMCID: PMC4289021
- DOI: 10.1038/nature13826
Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal
Abstract
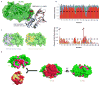
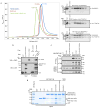
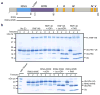
Protein poly(ADP-ribosyl)ation (PARylation) has a role in diverse cellular processes such as DNA repair, transcription, Wnt signalling, and cell death. Recent studies have shown that PARylation can serve as a signal for the polyubiquitination and degradation of several crucial regulatory proteins, including Axin and 3BP2 (refs 7, 8, 9). The RING-type E3 ubiquitin ligase RNF146 (also known as Iduna) is responsible for PARylation-dependent ubiquitination (PARdU). Here we provide a structural basis for RNF146-catalysed PARdU and how PARdU specificity is achieved. First, we show that iso-ADP-ribose (iso-ADPr), the smallest internal poly(ADP-ribose) (PAR) structural unit, binds between the WWE and RING domains of RNF146 and functions as an allosteric signal that switches the RING domain from a catalytically inactive state to an active one. In the absence of PAR, the RING domain is unable to bind and activate a ubiquitin-conjugating enzyme (E2) efficiently. Binding of PAR or iso-ADPr induces a major conformational change that creates a functional RING structure. Thus, RNF146 represents a new mechanistic class of RING E3 ligases, the activities of which are regulated by non-covalent ligand binding, and that may provide a template for designing inducible protein-degradation systems. Second, we find that RNF146 directly interacts with the PAR polymerase tankyrase (TNKS). Disruption of the RNF146-TNKS interaction inhibits turnover of the substrate Axin in cells. Thus, both substrate PARylation and PARdU are catalysed by enzymes within the same protein complex, and PARdU substrate specificity may be primarily determined by the substrate-TNKS interaction. We propose that the maintenance of unliganded RNF146 in an inactive state may serve to maintain the stability of the RNF146-TNKS complex, which in turn regulates the homeostasis of PARdU activity in the cell.
Conflict of interest statement
The authors declare no competing financial interests.
Figures










References
-
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. - PubMed
-
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. - PubMed
-
- Virag L. 50Years of poly(ADP-ribosyl)ation. Molecular aspects of medicine. 2013;34:1043–1045. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

