Solvent-Free, Highly Coarse-Grained Models for Charged Lipid Systems
- PMID: 25328498
- PMCID: PMC4196741
- DOI: 10.1021/ct500474a
Solvent-Free, Highly Coarse-Grained Models for Charged Lipid Systems
Abstract
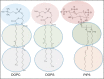
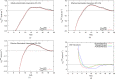
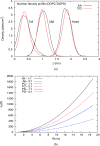
We present a methodology to develop coarse-grained lipid models such that electrostatic interactions between the coarse-grained sites can be derived accurately from an all-atom molecular dynamics trajectory and expressed as an effective pairwise electrostatic potential with appropriate screening functions. The reference nonbonded forces from the all-atom trajectory are decomposed into separate electrostatic and van der Waals (vdW) forces, based on the multiscale coarse-graining method. The coarse-grained electrostatic potential is assumed to be a general function of unknown variables and the final site-site interactions are obtained variationally, where the residual of the electrostatic forces from the assumed field is minimized. The resulting electrostatic interactions are fitted to screened electrostatics functions, with a special treatment for distance-dependent dielectrics and screened dipole-dipole interactions. The vdW interactions are derived separately. The resulting charged hybrid coarse-graining method is applied to various solvent-free three-site models of anionic lipid systems.
Figures



Similar articles
-
Conditional Reversible Work Coarse-Grained Models of Molecular Liquids with Coulomb Electrostatics - A Proof of Concept Study on Weakly Polar Organic Molecules.J Chem Theory Comput. 2017 Dec 12;13(12):6158-6166. doi: 10.1021/acs.jctc.7b00611. Epub 2017 Dec 1. J Chem Theory Comput. 2017. PMID: 29161026
-
A coarse-grained protein-protein potential derived from an all-atom force field.J Phys Chem B. 2007 Aug 9;111(31):9390-9. doi: 10.1021/jp0727190. Epub 2007 Jul 6. J Phys Chem B. 2007. PMID: 17616119
-
A Hybrid Approach for Highly Coarse-grained Lipid Bilayer Models.J Chem Theory Comput. 2013 Jan 8;9(1):750-765. doi: 10.1021/ct300751h. J Chem Theory Comput. 2013. PMID: 25100925 Free PMC article.
-
Adaptive resolution simulations of biomolecular systems.Eur Biophys J. 2017 Dec;46(8):821-835. doi: 10.1007/s00249-017-1248-0. Epub 2017 Sep 13. Eur Biophys J. 2017. PMID: 28905203 Review.
-
Coarse-graining methods for computational biology.Annu Rev Biophys. 2013;42:73-93. doi: 10.1146/annurev-biophys-083012-130348. Epub 2013 Feb 28. Annu Rev Biophys. 2013. PMID: 23451897 Review.
Cited by
-
Utilizing Machine Learning to Greatly Expand the Range and Accuracy of Bottom-Up Coarse-Grained Models through Virtual Particles.J Chem Theory Comput. 2023 Jul 25;19(14):4402-4413. doi: 10.1021/acs.jctc.2c01183. Epub 2023 Feb 20. J Chem Theory Comput. 2023. PMID: 36802592 Free PMC article.
-
Multiscale design of coarse-grained elastic network-based potentials for the μ opioid receptor.J Mol Model. 2016 Sep;22(9):227. doi: 10.1007/s00894-016-3092-z. Epub 2016 Aug 26. J Mol Model. 2016. PMID: 27566318
-
Biophysics of membrane curvature remodeling at molecular and mesoscopic lengthscales.J Phys Condens Matter. 2018 Jul 11;30(27):273001. doi: 10.1088/1361-648X/aac702. Epub 2018 May 22. J Phys Condens Matter. 2018. PMID: 29786613 Free PMC article.
-
Multiscale simulations of protein-facilitated membrane remodeling.J Struct Biol. 2016 Oct;196(1):57-63. doi: 10.1016/j.jsb.2016.06.012. Epub 2016 Jun 17. J Struct Biol. 2016. PMID: 27327264 Free PMC article. Review.
-
Computational Modeling of Realistic Cell Membranes.Chem Rev. 2019 May 8;119(9):6184-6226. doi: 10.1021/acs.chemrev.8b00460. Epub 2019 Jan 9. Chem Rev. 2019. PMID: 30623647 Free PMC article. Review.
References
-
- Alberts B.; Johnson A.; Lewis L.; Raff M.; Roberts K.; Walter P.. Molecular Biology of the Cell; Taylor and Francis: New York, 2002.
-
- Karp G.Cell and Molecular Biology: Concepts and Experiments; John Wiley & Sons: Toronto, Canada, 2007.
-
- Grecco H. E.; Schmick M.; Bastiaens P. I. H. Signaling from the living plasma membrane. Cell 2011, 144, 897–909. - PubMed
-
- Kusumi A.; Fujiwara T. K.; Chadda R.; Xie M.; Tsunoyama T. A.; Kalay Z.; Kasai R. S.; Suzuki K. G. N. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 2012, 28, 215–250. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
