A metagenomic study of the microbial communities in four parallel biogas reactors
- PMID: 25328537
- PMCID: PMC4200192
- DOI: 10.1186/s13068-014-0146-2
A metagenomic study of the microbial communities in four parallel biogas reactors
Abstract
Background: Biogas is a renewable energy carrier which is used for heat and power production or, in the form of purified methane, as a vehicle fuel. The formation of methane from organic materials is carried out by a mixed microbial community under anaerobic conditions. However, details about the microbes involved and their function are limited. In this study we compare the metagenomes of four parallel biogas reactors digesting a protein-rich substrate, relate microbiology to biogas performance, and observe differences in these reactors' microbial communities compared to the original inoculum culture.
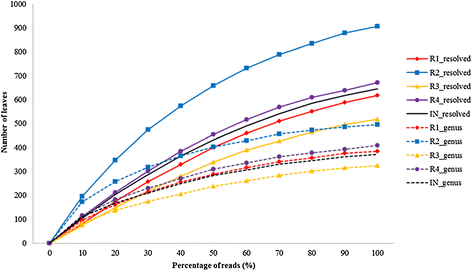
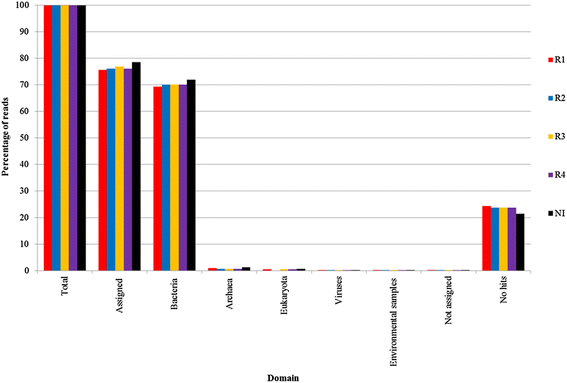
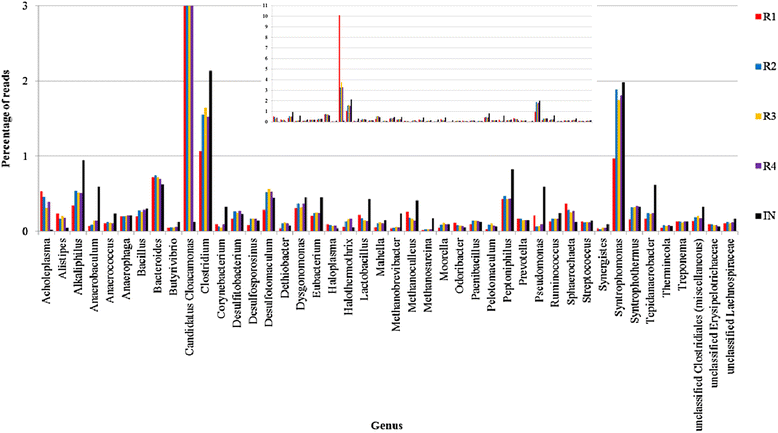
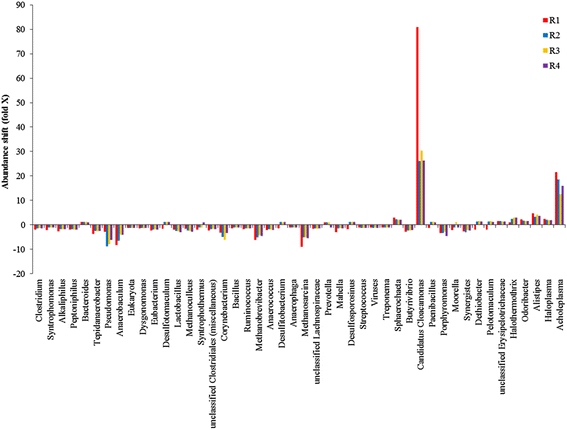
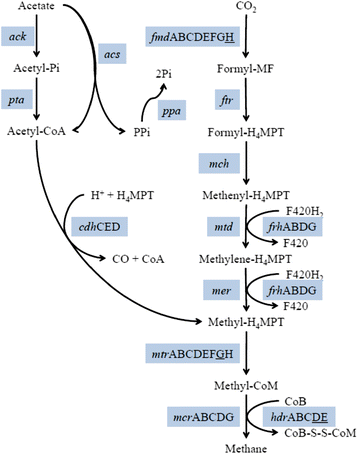
Results: The biogas process performance during the startup phase of four parallel continuous stirred tank reactors (designated R1, R2, R3, and R4) co-digesting fish waste and cow manure was studied. The microbial composition of the inoculum (day 0) and the four reactors at day 59 was studied and compared using 454 FLX Titanium pyrosequencing. In the inoculum and the reactor samples, the Bacteria Clostridium and Syntrophomonas were highly abundant, and the dominating methanogen was the hydrogenotrophic Methanoculleus. Syntrophic prokaryotes frequently found in biogas reactors with high concentrations of ammonium and volatile fatty acids were detected in all samples. The species Candidatus Cloacimonas acidaminovorans of the candidate phylum Cloacimonetes (WWE1) increased in all reactors and was the dominating bacterium at day 59. In particular, this bacterium showed a very high abundance in R1, which distinguished this reactor significantly from the other reactors in terms of microbial composition. Methane production and the reactor slurry characteristics were monitored in the digestion period. Generally all four reactors operated stably and showed rather similar characteristics. The average methane production in the reactors varied between 0.278 and 0.296 L gVS(-1), with the lowest production in R1.
Conclusions: This study showed that four parallel reactors co-digesting manure and fish waste silage operated stably during a startup phase. Several important Archaea and Bacteria degrading the protein-rich substrate were identified. In particular, microorganisms involved in syntrophic methane production seemed to be important. The detailed characterization of the microbial communities presented in this work may be useful for the operation of biogas plants degrading substrates with high concentrations of proteins.
Keywords: Anaerobic digestion; Biofuel; Biogas; Biorefinery; Metagenomic; Methane; Syntrophic oxidation; Taxonomic structure.
Figures





References
-
- Statistics Norway. SSB. 2013.Avfallsregnskapet 2011.http://www.ssb.no/natur-og-miljo/statistikker/avfregno.
-
- Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD, Lykidis A, Sieprawska-Lupa M, Dharmarajan L, Goltsman E, Lapidus A, Saunders E, Han C, Land M, Lucas S, Mukhopadhyay B, Whitman WB, Woese C, Bristow J, Kyrpides N. Genomic characterization of methanomicrobiales reveals three classes of methanogens. PLoS One. 2009;4:6. - PMC - PubMed
-
- Zinder SH. Physiological ecology of methanogenesis. In: Ferry JG, editor. Methanogenesis. Ecology, Physiology, Biochemistry and Genetics. Volume 1. 1. New York: Chapman and Hall; 1993. pp. 128–206.
LinkOut - more resources
Full Text Sources
Other Literature Sources

