Hydrophobicity and aromaticity are primary factors shaping variation in amino acid usage of chicken proteome
- PMID: 25329059
- PMCID: PMC4199684
- DOI: 10.1371/journal.pone.0110381
Hydrophobicity and aromaticity are primary factors shaping variation in amino acid usage of chicken proteome
Abstract
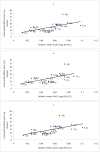
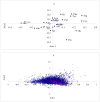
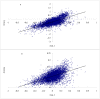
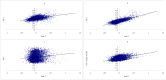
Amino acids are utilized with different frequencies both among species and among genes within the same genome. Up to date, no study on the amino acid usage pattern of chicken has been performed. In the present study, we carried out a systematic examination of the amino acid usage in the chicken proteome. Our data indicated that the relative amino acid usage is positively correlated with the tRNA gene copy number. GC contents, including GC1, GC2, GC3, GC content of CDS and GC content of the introns, were correlated with the most of the amino acid usage, especially for GC rich and GC poor amino acids, however, multiple linear regression analyses indicated that only approximately 10-40% variation of amino acid usage can be explained by GC content for GC rich and GC poor amino acids. For other intermediate GC content amino acids, only approximately 10% variation can be explained. Correspondence analyses demonstrated that the main factors responsible for the variation of amino acid usage in chicken are hydrophobicity, aromaticity and genomic GC content. Gene expression level also influenced the amino acid usage significantly. We argued that the amino acid usage of chicken proteome likely reflects a balance or near balance between the action of selection, mutation, and genetic drift.
Conflict of interest statement
Figures
References
-
- Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol 44: 282–288. - PubMed
-
- Duret L (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet 16: 287–289. - PubMed
-
- Kanaya S, Yamada Y, Kudo Y, Ikemura T (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143–155. - PubMed
-
- Novoa EM, Ribas de Pouplana L (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28(11): 574–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

