Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines
- PMID: 25329162
- PMCID: PMC4199505
- DOI: 10.1371/journal.pgen.1004720
Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines
Abstract
Hsp100 family chaperones of microorganisms and plants cooperate with the Hsp70/Hsp40/NEF system to resolubilize and reactivate stress-denatured proteins. In yeast this machinery also promotes propagation of prions by fragmenting prion polymers. We previously showed the bacterial Hsp100 machinery cooperates with the yeast Hsp40 Ydj1 to support yeast thermotolerance and with the yeast Hsp40 Sis1 to propagate [PSI+] prions. Here we find these Hsp40s similarly directed specific activities of the yeast Hsp104-based machinery. By assessing the ability of Ydj1-Sis1 hybrid proteins to complement Ydj1 and Sis1 functions we show their C-terminal substrate-binding domains determined distinctions in these and other cellular functions of Ydj1 and Sis1. We find propagation of [URE3] prions was acutely sensitive to alterations in Sis1 activity, while that of [PIN+] prions was less sensitive than [URE3], but more sensitive than [PSI+]. These findings support the ideas that overexpressing Ydj1 cures [URE3] by competing with Sis1 for interaction with the Hsp104-based disaggregation machine, and that different prions rely differently on activity of this machinery, which can explain the various ways they respond to alterations in chaperone function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
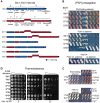


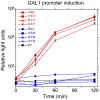


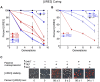

References
-
- Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. - PubMed
-
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, et al. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665. - PubMed
-
- Lum R, Tkach JM, Vierling E, Glover JR (2004) Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146. - PubMed
-
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

