Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway
- PMID: 25329237
- PMCID: PMC4755588
- DOI: 10.1111/1462-2920.12669
Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway
Abstract
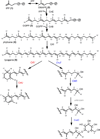
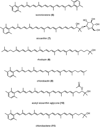
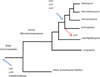
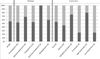
Members of the marine actinomycete genus Salinispora constitutively produce a characteristic orange pigment during vegetative growth. Contrary to the understanding of widespread carotenoid biosynthesis pathways in bacteria, Salinispora carotenoid biosynthesis genes are not confined to a single cluster. Instead, bioinformatic and genetic investigations confirm that four regions of the Salinispora tropica CNB-440 genome, consisting of two gene clusters and two independent genes, contribute to the in vivo production of a single carotenoid. This compound, namely (2'S)-1'-(β-D-glucopyranosyloxy)-3',4'-didehydro-1',2'-dihydro-φ,ψ-caroten-2'-ol, is novel and has been given the trivial name 'sioxanthin'. Sioxanthin is a C40 -carotenoid, glycosylated on one end of the molecule and containing an aryl moiety on the opposite end. Glycosylation is unusual among actinomycete carotenoids, and sioxanthin joins a rare group of carotenoids with polar and non-polar head groups. Gene sequence homology predicts that the sioxanthin biosynthetic pathway is present in all of the Salinispora as well as other members of the family Micromonosporaceae. Additionally, this study's investigations of clustering of carotenoid biosynthetic genes in heterotrophic bacteria show that a non-clustered genome arrangement is more common than previously suggested, with nearly half of the investigated genomes showing a non-clustered architecture.
© 2014 Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures

References
-
- Ahmed L, Jensen PR, Freel KC, Brown R, Jones AL, Kim B-Y, Goodfellow M. Salinispora pacifica sp nov., an actinomycete from marine sediments. Anton Leeuw Int J G. 2013;103:1069–1078. - PubMed
-
- Armstrong GA. Genetics of eubacterial carotenoid biosynthesis: A colorful tale. In: Ornston LN, editor. Annu Rev Microbiol. USA: Annual Reviews Inc.; 1997. pp. 629–659. - PubMed
-
- Armstrong GA, Hearst JE. Carotenoids 2: Genetics and molecular biology of carotenoid pigment biosynthesis. FASEB J. 1996;10:228–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

