Diversity analysis in Cannabis sativa based on large-scale development of expressed sequence tag-derived simple sequence repeat markers
- PMID: 25329551
- PMCID: PMC4203809
- DOI: 10.1371/journal.pone.0110638
Diversity analysis in Cannabis sativa based on large-scale development of expressed sequence tag-derived simple sequence repeat markers
Abstract
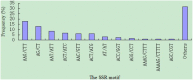
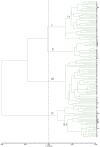
Cannabis sativa L. is an important economic plant for the production of food, fiber, oils, and intoxicants. However, lack of sufficient simple sequence repeat (SSR) markers has limited the development of cannabis genetic research. Here, large-scale development of expressed sequence tag simple sequence repeat (EST-SSR) markers was performed to obtain more informative genetic markers, and to assess genetic diversity in cannabis (Cannabis sativa L.). Based on the cannabis transcriptome, 4,577 SSRs were identified from 3,624 ESTs. From there, a total of 3,442 complementary primer pairs were designed as SSR markers. Among these markers, trinucleotide repeat motifs (50.99%) were the most abundant, followed by hexanucleotide (25.13%), dinucleotide (16.34%), tetranucloetide (3.8%), and pentanucleotide (3.74%) repeat motifs, respectively. The AAG/CTT trinucleotide repeat (17.96%) was the most abundant motif detected in the SSRs. One hundred and seventeen EST-SSR markers were randomly selected to evaluate primer quality in 24 cannabis varieties. Among these 117 markers, 108 (92.31%) were successfully amplified and 87 (74.36%) were polymorphic. Forty-five polymorphic primer pairs were selected to evaluate genetic diversity and relatedness among the 115 cannabis genotypes. The results showed that 115 varieties could be divided into 4 groups primarily based on geography: Northern China, Europe, Central China, and Southern China. Moreover, the coefficient of similarity when comparing cannabis from Northern China with the European group cannabis was higher than that when comparing with cannabis from the other two groups, owing to a similar climate. This study outlines the first large-scale development of SSR markers for cannabis. These data may serve as a foundation for the development of genetic linkage, quantitative trait loci mapping, and marker-assisted breeding of cannabis.
Conflict of interest statement
Figures

Similar articles
-
Development and characterization of 1,827 expressed sequence tag-derived simple sequence repeat markers for ramie (Boehmeria nivea L. Gaud).PLoS One. 2013;8(4):e60346. doi: 10.1371/journal.pone.0060346. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565230 Free PMC article.
-
Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.).Mol Biol Rep. 2020 Dec;47(12):9385-9397. doi: 10.1007/s11033-020-05981-8. Epub 2020 Nov 19. Mol Biol Rep. 2020. PMID: 33215363
-
Characterization and development of EST-SSR markers to study the genetic diversity and populations analysis of Jerusalem artichoke (Helianthus tuberosus L.).Genes Genomics. 2018 Oct;40(10):1023-1032. doi: 10.1007/s13258-018-0708-y. Epub 2018 Jun 28. Genes Genomics. 2018. PMID: 29956221
-
Exploitation of pepper EST-SSRs and an SSR-based linkage map.Theor Appl Genet. 2006 Dec;114(1):113-30. doi: 10.1007/s00122-006-0415-y. Epub 2006 Oct 18. Theor Appl Genet. 2006. PMID: 17047912
-
Potentials and Challenges of Genomics for Breeding Cannabis Cultivars.Front Plant Sci. 2020 Sep 25;11:573299. doi: 10.3389/fpls.2020.573299. eCollection 2020. Front Plant Sci. 2020. PMID: 33101342 Free PMC article. Review.
Cited by
-
Genome-wide development of insertion-deletion (InDel) markers for Cannabis and its uses in genetic structure analysis of Chinese germplasm and sex-linked marker identification.BMC Genomics. 2021 Aug 5;22(1):595. doi: 10.1186/s12864-021-07883-w. BMC Genomics. 2021. PMID: 34353285 Free PMC article.
-
Development of SSR markers for genetic diversity analysis and species identification in Polygonatum odoratum (Mill.) Druce based on transcriptome sequences.PLoS One. 2024 Sep 23;19(9):e0308316. doi: 10.1371/journal.pone.0308316. eCollection 2024. PLoS One. 2024. PMID: 39312515 Free PMC article.
-
Developing and Testing Molecular Markers in Cannabis sativa (Hemp) for Their Use in Variety and Dioecy Assessments.Plants (Basel). 2021 Oct 14;10(10):2174. doi: 10.3390/plants10102174. Plants (Basel). 2021. PMID: 34685983 Free PMC article.
-
Transcriptome analysis and molecular marker discovery in Solanum incanum and S. aethiopicum, two close relatives of the common eggplant (Solanum melongena) with interest for breeding.BMC Genomics. 2016 Apr 23;17:300. doi: 10.1186/s12864-016-2631-4. BMC Genomics. 2016. PMID: 27108408 Free PMC article.
-
Comparative Genetic Structure of Cannabis sativa Including Federally Produced, Wild Collected, and Cultivated Samples.Front Plant Sci. 2021 Sep 29;12:675770. doi: 10.3389/fpls.2021.675770. eCollection 2021. Front Plant Sci. 2021. PMID: 34707624 Free PMC article.
References
-
- Gilmore S, Peakall R (2003) Isolation of microsatellite markers in Cannabis sativa L. (marijuana). Mol Ecol Notes 3: 105–107.
-
- Alghanim HJ, Almirall JR (2003) Development of microsatellite markers in Cannabis sativa for DNA typing and genetic relatedness analyses. Anal Bioanal Chem 376: 1225–1233. - PubMed
-
- Gillan R, Cole MD, Linacre A, Thorpe JW, Watson ND (1995) Comparison of Cannabis sativa by random amplification of polymorphic DNA (RAPD) and HPLC of cannabinoid: a preliminary study. Sci Justice 35: 169–177. - PubMed
-
- Jagadish V, Robertson J, Gibbs A (1996) RAPD analysis distinguished Cannabis sativa samples from different sources. Foren Sci Int 79: 113–121.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

