How structure defines affinity in protein-protein interactions
- PMID: 25329579
- PMCID: PMC4199723
- DOI: 10.1371/journal.pone.0110085
How structure defines affinity in protein-protein interactions
Abstract
Protein-protein interactions (PPI) in nature are conveyed by a multitude of binding modes involving various surfaces, secondary structure elements and intermolecular interactions. This diversity results in PPI binding affinities that span more than nine orders of magnitude. Several early studies attempted to correlate PPI binding affinities to various structure-derived features with limited success. The growing number of high-resolution structures, the appearance of more precise methods for measuring binding affinities and the development of new computational algorithms enable more thorough investigations in this direction. Here, we use a large dataset of PPI structures with the documented binding affinities to calculate a number of structure-based features that could potentially define binding energetics. We explore how well each calculated biophysical feature alone correlates with binding affinity and determine the features that could be used to distinguish between high-, medium- and low- affinity PPIs. Furthermore, we test how various combinations of features could be applied to predict binding affinity and observe a slow improvement in correlation as more features are incorporated into the equation. In addition, we observe a considerable improvement in predictions if we exclude from our analysis low-resolution and NMR structures, revealing the importance of capturing exact intermolecular interactions in our calculations. Our analysis should facilitate prediction of new interactions on the genome scale, better characterization of signaling networks and design of novel binding partners for various target proteins.
Conflict of interest statement
Figures

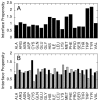
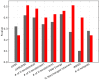
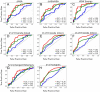

References
-
- Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, et al. (1997) Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature 387: 630–634. - PubMed
-
- Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, et al. (1996) CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 384: 577–581. - PubMed
-
- Kobe B, Deisenhofer J (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374: 183–186. - PubMed
-
- Vicentini AM, Kieffer B, Matthies R, Meyhack B, Hemmings BA, et al. (1990) Protein chemical and kinetic characterization of recombinant porcine ribonuclease inhibitor expressed in Saccharomyces cerevisiae. Biochemistry 29: 8827–8834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

