Suppression of Foxo1 activity and down-modulation of CD62L (L-selectin) in HIV-1 infected resting CD4 T cells
- PMID: 25330112
- PMCID: PMC4199762
- DOI: 10.1371/journal.pone.0110719
Suppression of Foxo1 activity and down-modulation of CD62L (L-selectin) in HIV-1 infected resting CD4 T cells
Abstract
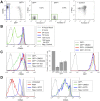
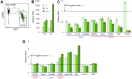
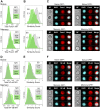
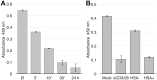
HIV-1 hijacks and disrupts many processes in the cells it infects in order to suppress antiviral immunity and to facilitate its replication. Resting CD4 T cells are important early targets of HIV-1 infection in which HIV-1 must overcome intrinsic barriers to viral replication. Although resting CD4 T cells are refractory to infection in vitro, local environmental factors within lymphoid and mucosal tissues such as cytokines facilitate viral replication while maintaining the resting state. These factors can be utilized in vitro to study HIV-1 replication in resting CD4 T cells. In vivo, the migration of resting naïve and central memory T cells into lymphoid tissues is dependent upon expression of CD62L (L-selectin), a receptor that is subsequently down-modulated following T cell activation. CD62L gene transcription is maintained in resting T cells by Foxo1 and KLF2, transcription factors that maintain T cell quiescence and which regulate additional cellular processes including survival, migration, and differentiation. Here we report that HIV-1 down-modulates CD62L in productively infected naïve and memory resting CD4 T cells while suppressing Foxo1 activity and the expression of KLF2 mRNA. Partial T cell activation was further evident as an increase in CD69 expression. Several other Foxo1- and KLF2-regulated mRNA were increased or decreased in productively infected CD4 T cells, including IL-7rα, Myc, CCR5, Fam65b, S1P1 (EDG1), CD52, Cyclin D2 and p21CIP1, indicating a profound reprogramming of these cells. The Foxo1 inhibitor AS1842856 accelerated de novo viral gene expression and the sequella of infection, supporting the notion that HIV-1 suppression of Foxo1 activity may be a strategy to promote replication in resting CD4 T cells. As Foxo1 is an investigative cancer therapy target, the development of Foxo1 interventions may assist the quest to specifically suppress or activate HIV-1 replication in vivo.
Conflict of interest statement
Figures


References
-
- Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, et al. (1999) Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286: 1353–1357. - PubMed
-
- Schacker T, Little S, Connick E, Gebhard K, Zhang ZQ, et al. (2001) Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis 183: 555–562. - PubMed
-
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, et al. (2005) Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434: 1148–1152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

