Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis
- PMID: 25330213
- PMCID: PMC4199496
- DOI: 10.1371/journal.pgen.1004664
Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis
Abstract
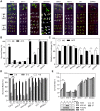
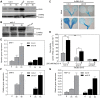
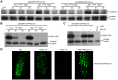
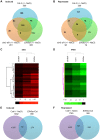
Ethylene has been regarded as a stress hormone to regulate myriad stress responses. Salinity stress is one of the most serious abiotic stresses limiting plant growth and development. But how ethylene signaling is involved in plant response to salt stress is poorly understood. Here we showed that Arabidopsis plants pretreated with ethylene exhibited enhanced tolerance to salt stress. Gain- and loss-of-function studies demonstrated that EIN3 (ETHYLENE INSENSITIVE 3) and EIL1 (EIN3-LIKE 1), two ethylene-activated transcription factors, are necessary and sufficient for the enhanced salt tolerance. High salinity induced the accumulation of EIN3/EIL1 proteins by promoting the proteasomal degradation of two EIN3/EIL1-targeting F-box proteins, EBF1 and EBF2, in an EIN2-independent manner. Whole-genome transcriptome analysis identified a list of SIED (Salt-Induced and EIN3/EIL1-Dependent) genes that participate in salt stress responses, including several genes encoding reactive oxygen species (ROS) scavengers. We performed a genetic screen for ein3 eil1-like salt-hypersensitive mutants and identified 5 EIN3 direct target genes including a previously unknown gene, SIED1 (At5g22270), which encodes a 93-amino acid polypeptide involved in ROS dismissal. We also found that activation of EIN3 increased peroxidase (POD) activity through the direct transcriptional regulation of PODs expression. Accordingly, ethylene pretreatment or EIN3 activation was able to preclude excess ROS accumulation and increased tolerance to salt stress. Taken together, our study provides new insights into the molecular action of ethylene signaling to enhance plant salt tolerance, and elucidates the transcriptional network of EIN3 in salt stress response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis.Plant Cell. 2010 Jul;22(7):2384-401. doi: 10.1105/tpc.110.076588. Epub 2010 Jul 20. Plant Cell. 2010. PMID: 20647342 Free PMC article.
-
The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling.Plant Cell. 2007 Feb;19(2):509-23. doi: 10.1105/tpc.106.048140. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307926 Free PMC article.
-
An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis.Plant Physiol. 2011 Oct;157(2):854-65. doi: 10.1104/pp.111.179028. Epub 2011 Aug 10. Plant Physiol. 2011. PMID: 21832142 Free PMC article.
-
Ethylene signaling: new levels of complexity and regulation.Curr Opin Plant Biol. 2008 Oct;11(5):479-85. doi: 10.1016/j.pbi.2008.06.011. Epub 2008 Aug 7. Curr Opin Plant Biol. 2008. PMID: 18692429 Free PMC article. Review.
-
Paradigms and paradox in the ethylene signaling pathway and interaction network.Mol Plant. 2011 Jul;4(4):626-34. doi: 10.1093/mp/ssr042. Epub 2011 Jun 20. Mol Plant. 2011. PMID: 21690206 Review.
Cited by
-
Chemical induction of leaf senescence and powdery mildew resistance involves ethylene-mediated chlorophyll degradation and ROS metabolism in cucumber.Hortic Res. 2022 May 17;9:uhac101. doi: 10.1093/hr/uhac101. eCollection 2022. Hortic Res. 2022. PMID: 35795391 Free PMC article.
-
Analysis of Phytohormone Signal Transduction in Sophora alopecuroides under Salt Stress.Int J Mol Sci. 2021 Jul 7;22(14):7313. doi: 10.3390/ijms22147313. Int J Mol Sci. 2021. PMID: 34298928 Free PMC article.
-
Ethylene and Abscisic Acid Signaling Pathways Differentially Influence Tomato Resistance to Combined Powdery Mildew and Salt Stress.Front Plant Sci. 2017 Jan 9;7:2009. doi: 10.3389/fpls.2016.02009. eCollection 2016. Front Plant Sci. 2017. PMID: 28119708 Free PMC article.
-
Application of nanoparticles for salinity stress management and biofortification in wheat: a review of dual approaches and insights.Front Plant Sci. 2025 Jul 4;16:1592866. doi: 10.3389/fpls.2025.1592866. eCollection 2025. Front Plant Sci. 2025. PMID: 40688693 Free PMC article. Review.
-
The combination of linkage mapping, genome-wide association study, and dynamic transcriptome analysis reveals conserved candidate genes for salt tolerance in maize.Theor Appl Genet. 2025 Jul 19;138(8):186. doi: 10.1007/s00122-025-04975-z. Theor Appl Genet. 2025. PMID: 40684070
References
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258. - PubMed
-
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18. - PubMed
-
- Ecker JR (1995) The ethylene signal transduction pathway in plants. Science 268: 667–675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

