Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis
- PMID: 25330732
- PMCID: PMC4209041
- DOI: 10.1186/s12870-014-0271-x
Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis
Abstract
Background: MicroRNAs (miRNAs) are approximately 19 ~ 21 nucleotide noncoding RNAs produced by Dicer-catalyzed excision from stem-loop precursors. Many plant miRNAs have critical functions in development, nutrient homeostasis, abiotic stress responses, and pathogen responses via interaction with specific target mRNAs. Camellia sinensis is one of the most important commercial beverage crops in the world. However, miRNAs associated with cold stress tolerance in C. sinensis remains unexplored. The use of high-throughput sequencing can provide a much deeper understanding of miRNAs. To obtain more insight into the function of miRNAs in cold stress tolerance, Illumina sequencing of C. sinensis sRNA was conducted.
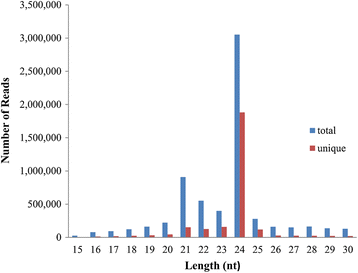
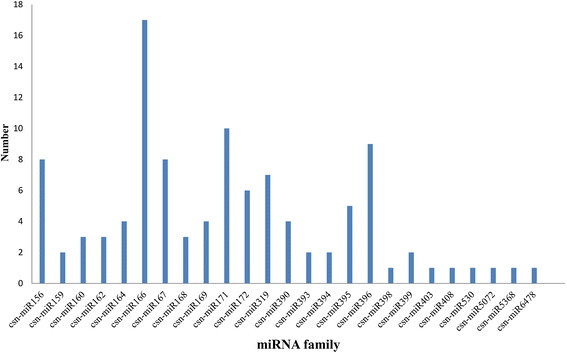
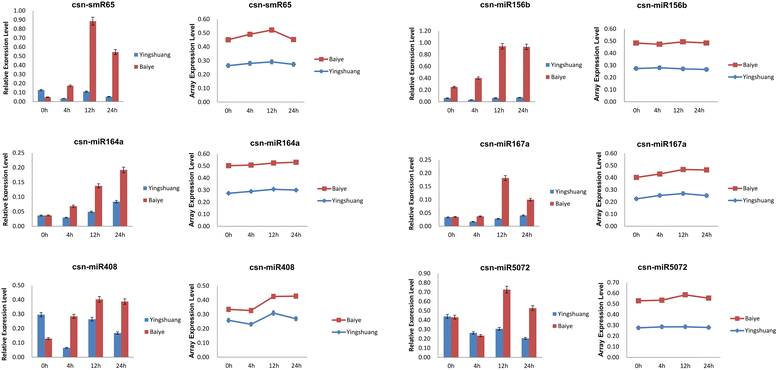
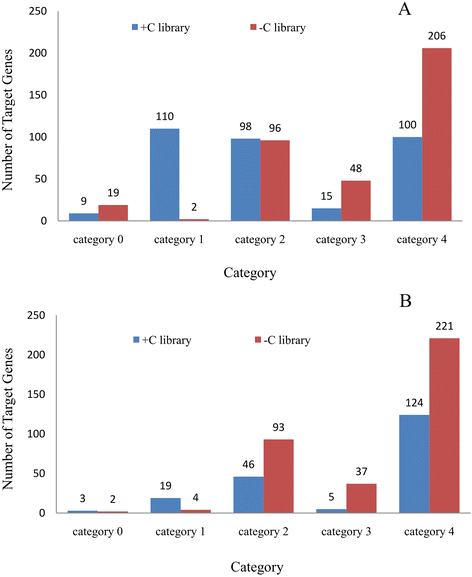
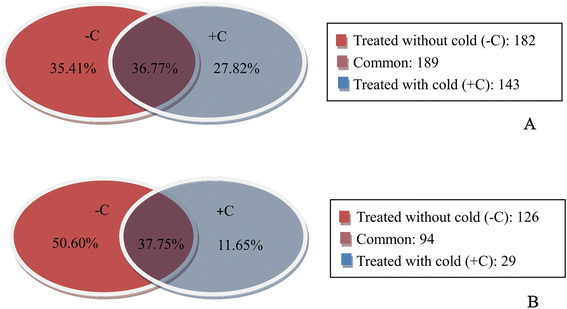
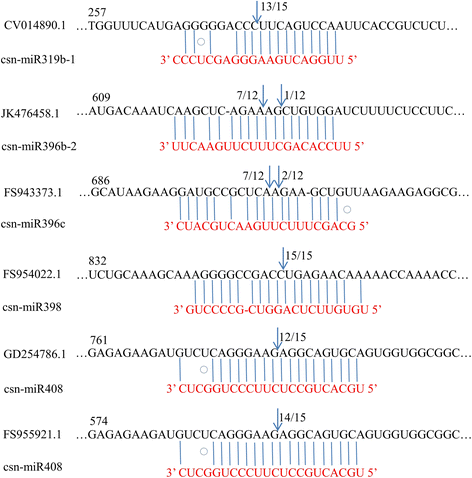
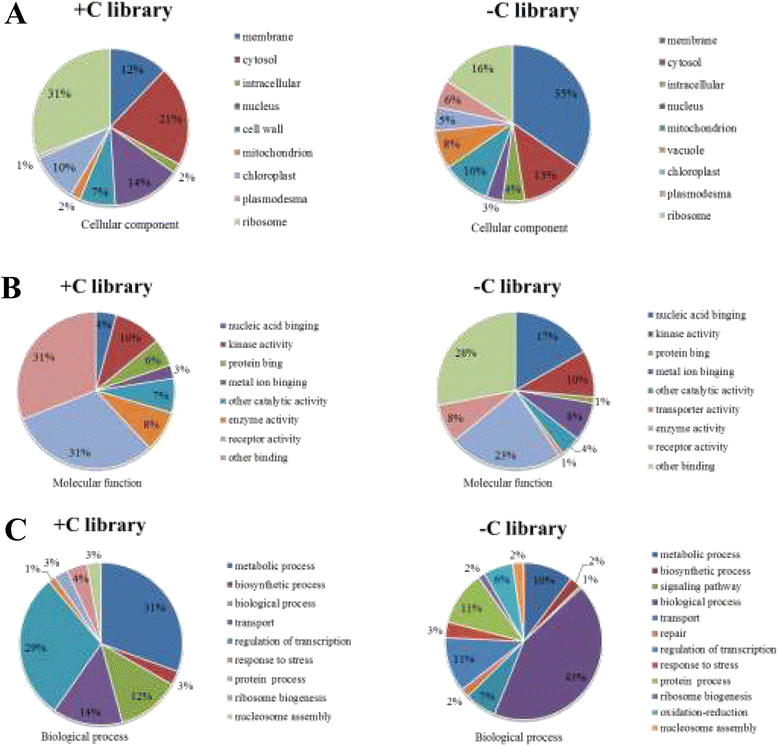
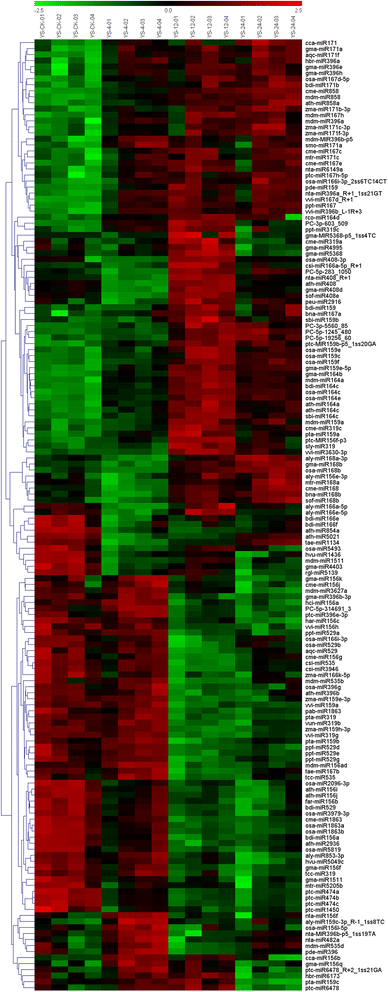
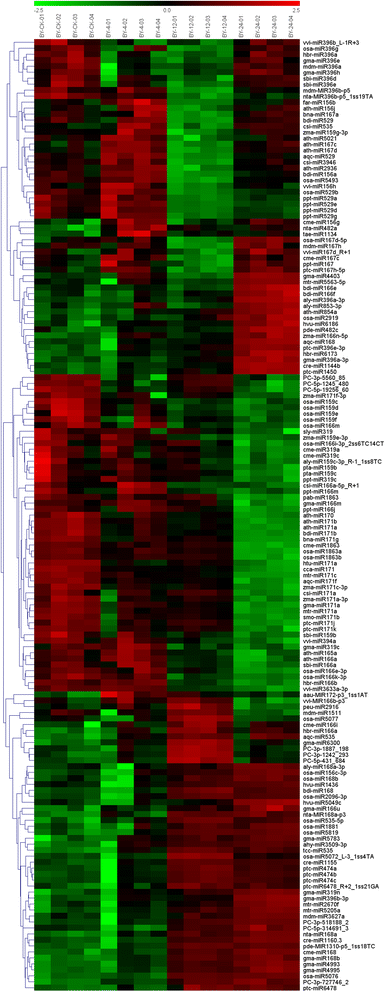
Result: Solexa sequencing technology was used for high-throughput sequencing of the small RNA library from the cold treatment of tea leaves. To align the sequencing data with known plant miRNAs, we characterized 106 conserved C. sinensis miRNAs. In addition, 215 potential candidate miRNAs were found, among, which 98 candidates with star sequences were chosen as novel miRNAs. Both congruously and differentially regulated miRNAs were obtained, and cultivar-specific miRNAs were identified by microarray-based hybridization in response to cold stress. The results were also confirmed by quantitative real-time polymerase chain reaction. To confirm the targets of miRNAs, two degradome libraries from two treatments were constructed. According to degradome sequencing, 455 and 591 genes were identified as cleavage targets of miRNAs from cold treatments and control libraries, respectively, and 283 targets were present in both libraries. Functional analysis of these miRNA targets indicated their involvement in important activities, such as development, regulation of transcription, and stress response.
Conclusions: We discovered 31 up-regulated miRNAs and 43 down-regulated miRNAs in 'Yingshuang', and 46 up-regulated miRNA and 45 down-regulated miRNAs in 'Baiye 1' in response to cold stress, respectively. A total of 763 related target genes were detected by degradome sequencing. The RLM-5'RACE procedure was successfully used to map the cleavage sites in six target genes of C. sinensis. These findings reveal important information about the regulatory mechanism of miRNAs in C. sinensis, and promote the understanding of miRNA functions during the cold response. The miRNA genotype-specific expression model might explain the distinct cold sensitivities between tea lines.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

