RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells
- PMID: 25330849
- PMCID: PMC4218956
- DOI: 10.1038/ncomms6220
RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells
Abstract
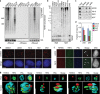
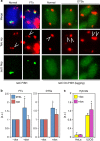
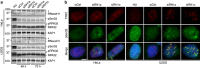
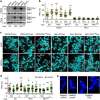
A fraction of cancer cells maintain telomeres through the telomerase-independent, 'Alternative Lengthening of Telomeres' (ALT) pathway. ALT relies on homologous recombination (HR) between telomeric sequences; yet, what makes ALT telomeres recombinogenic remains unclear. Here we show that the RNA endonuclease RNaseH1 regulates the levels of RNA-DNA hybrids between telomeric DNA and the long noncoding RNA TERRA, and is a key mediator of telomere maintenance in ALT cells. RNaseH1 associated to telomeres specifically in ALT cells and its depletion led to telomeric hybrid accumulation, exposure of single-stranded telomeric DNA, activation of replication protein A at telomeres and abrupt telomere excision. Conversely, overexpression of RNaseH1 weakened the recombinogenic nature of ALT telomeres and led to telomere shortening. Altering cellular RNaseH1 levels did not perturb telomere homoeostasis in telomerase-positive cells. RNaseH1 maintains regulated levels of telomeric RNA-DNA hybrids at ALT telomeres to trigger HR without compromising telomere integrity too severely.
Figures

References
-
- Palm W. & de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008). - PubMed
-
- Hug N. & Lingner J. Telomere length homoeostasis. Chromosoma 115, 413–425 (2006). - PubMed
-
- Arora R., Brun C. M. & Azzalin C. M. TERRA: long noncoding RNA at eukaryotic telomeres. Prog. Mol. Subcell. Biol. 51, 65–94 (2011). - PubMed
-
- Feuerhahn S., Iglesias N., Panza A., Porro A. & Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 584, 3812–3818 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

