A thyroid hormone receptor/KLF9 axis in human hepatocytes and pluripotent stem cells
- PMID: 25330987
- PMCID: PMC6317531
- DOI: 10.1002/stem.1875
A thyroid hormone receptor/KLF9 axis in human hepatocytes and pluripotent stem cells
Abstract
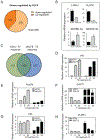
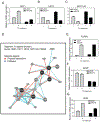
Biological processes require close cooperation of multiple transcription factors that integrate different signals. Thyroid hormone receptors (TRs) induce Krüppel-like factor 9 (KLF9) to regulate neurogenesis. Here, we show that triiodothyronine (T3) also works through TR to induce KLF9 in HepG2 liver cells, mouse liver, and mouse and human primary hepatocytes and sought to understand TR/KLF9 network function in the hepatocyte lineage and stem cells. Knockdown experiments reveal that KLF9 regulates hundreds of HepG2 target genes and modulates T3 response. Together, T3 and KLF9 target genes influence pathways implicated in stem cell self-renewal and differentiation, including Notch signaling, and we verify that T3 and KLF9 cooperate to regulate key Notch pathway genes and work independently to regulate others. T3 also induces KLF9 in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSC) and this effect persists during differentiation to definitive endoderm and hiPSC-derived hepatocytes. Microarray analysis reveals that T3 regulates hundreds of hESC and hiPSC target genes that cluster into many of the same pathways implicated in TR and KLF9 regulation in HepG2 cells. KLF9 knockdown confirms that TR and KLF9 cooperate to regulate Notch pathway genes in hESC and hiPSC, albeit in a partly cell-specific manner. Broader analysis of T3 responsive hESC/hiPSC genes suggests that TRs regulate multiple early steps in ESC differentiation. We propose that TRs cooperate with KLF9 to regulate hepatocyte proliferation and differentiation and early stages of organogenesis and that TRs exert widespread and important influences on ESC biology.
Keywords: Human embryonic stem cell; Induced pluripotent stem cell; Krüppel-like factor 9; Notch; Thyroid receptor.
© 2014 AlphaMed Press.
Conflict of interest statement
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Figures


References
-
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev 2001;81:1097–1142. - PubMed
-
- Hoopfer ED, Huang L, Denver RJ. Basic transcription element binding protein is a thyroid hormone-regulated transcription factor expressed during metamorphosis in Xenopus laevis. Dev Growth Differ 2002;44: 365–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

