Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes
- PMID: 25331115
- PMCID: PMC4263981
- DOI: 10.1182/blood-2014-06-582700
Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes
Abstract
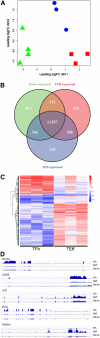
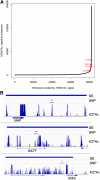
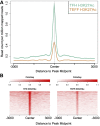
T follicular helper (Tfh) cells are a subset of CD4(+) T helper cells that migrate into germinal centers and promote B-cell maturation into memory B and plasma cells. Tfh cells are necessary for promotion of protective humoral immunity following pathogen challenge, but when aberrantly regulated, drive pathogenic antibody formation in autoimmunity and undergo neoplastic transformation in angioimmunoblastic T-cell lymphoma and other primary cutaneous T-cell lymphomas. Limited information is available on the expression and regulation of genes in human Tfh cells. Using a fluorescence-activated cell sorting-based strategy, we obtained primary Tfh and non-Tfh T effector cells from tonsils and prepared genome-wide maps of active, intermediate, and poised enhancers determined by chromatin immunoprecipitation-sequencing, with parallel transcriptome analyses determined by RNA sequencing. Tfh cell enhancers were enriched near genes highly expressed in lymphoid cells or involved in lymphoid cell function, with many mapping to sites previously associated with autoimmune disease in genome-wide association studies. A group of active enhancers unique to Tfh cells associated with differentially expressed genes was identified. Fragments from these regions directed expression in reporter gene assays. These data provide a significant resource for studies of T lymphocyte development and differentiation and normal and perturbed Tfh cell function.
© 2014 by The American Society of Hematology.
Figures


References
-
- Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol. 2011;29:621–663. - PubMed
-
- Weinmann AS. Regulatory mechanisms that control T-follicular helper and T-helper 1 cell flexibility. Immunol Cell Biol. 2014;92(1):34–39. - PubMed
-
- Crotty S. The 1-1-1 fallacy. Immunol Rev. 2012;247(1):133–142. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- R21AR062842/AR/NIAMS NIH HHS/United States
- R01 HL106184/HL/NHLBI NIH HHS/United States
- R37 AR040072/AR/NIAMS NIH HHS/United States
- T32HD007094/HD/NICHD NIH HHS/United States
- R01 AR040072/AR/NIAMS NIH HHS/United States
- T32 HD007094/HD/NICHD NIH HHS/United States
- R21 AR062842/AR/NIAMS NIH HHS/United States
- R01HL106184/HL/NHLBI NIH HHS/United States
- P30AR053495/AR/NIAMS NIH HHS/United States
- R01 HL065448/HL/NHLBI NIH HHS/United States
- R01HL65448/HL/NHLBI NIH HHS/United States
- T32AR07107/AR/NIAMS NIH HHS/United States
- T32 AR007107/AR/NIAMS NIH HHS/United States
- R01AR040072/AR/NIAMS NIH HHS/United States
- P30 AR053495/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

