N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity
- PMID: 25332400
- PMCID: PMC4227797
- DOI: 10.1093/nar/gku995
N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity
Abstract
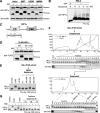
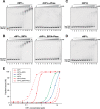
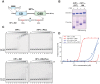
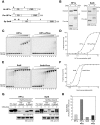
Heterochromatin protein 1 (HP1) is an evolutionarily conserved chromosomal protein that binds to lysine 9-methylated histone H3 (H3K9me), a hallmark of heterochromatin. Although HP1 phosphorylation has been described in several organisms, the biological implications of this modification remain largely elusive. Here we show that HP1's phosphorylation has a critical effect on its nucleosome binding properties. By in vitro phosphorylation assays and conventional chromatography, we demonstrated that casein kinase II (CK2) is the kinase primarily responsible for phosphorylating the N-terminus of human HP1α. Pull-down assays using in vitro-reconstituted nucleosomes showed that unmodified HP1α bound H3K9-methylated and H3K9-unmethylated nucleosomes with comparable affinity, whereas CK2-phosphorylated HP1α showed a high specificity for H3K9me3-modified nucleosomes. Electrophoretic mobility shift assays showed that CK2-mediated phosphorylation diminished HP1α's intrinsic DNA binding, which contributed to its H3K9me-independent nucleosome binding. CK2-mediated phosphorylation had a similar effect on the nucleosome-binding specificity of fly HP1a and S. pombe Swi6. These results suggested that HP1 phosphorylation has an evolutionarily conserved role in HP1's recognition of H3K9me-marked nucleosomes.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Grewal S.I., Elgin S.C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. - PubMed
-
- Richards E.J., Elgin S.C. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. - PubMed
-
- Grewal S.I., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

