Removing batch effects for prediction problems with frozen surrogate variable analysis
- PMID: 25332844
- PMCID: PMC4179553
- DOI: 10.7717/peerj.561
Removing batch effects for prediction problems with frozen surrogate variable analysis
Abstract
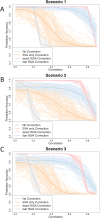
Batch effects are responsible for the failure of promising genomic prognostic signatures, major ambiguities in published genomic results, and retractions of widely-publicized findings. Batch effect corrections have been developed to remove these artifacts, but they are designed to be used in population studies. But genomic technologies are beginning to be used in clinical applications where samples are analyzed one at a time for diagnostic, prognostic, and predictive applications. There are currently no batch correction methods that have been developed specifically for prediction. In this paper, we propose an new method called frozen surrogate variable analysis (fSVA) that borrows strength from a training set for individual sample batch correction. We show that fSVA improves prediction accuracy in simulations and in public genomic studies. fSVA is available as part of the sva Bioconductor package.
Keywords: Batch effects; Database; Genomics; Machine learning; Prediction; Statistics; Surrogate variable analysis.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

