Characterization of the highly variable immune response gene family, He185/333, in the sea urchin, Heliocidaris erythrogramma
- PMID: 25333281
- PMCID: PMC4204807
- DOI: 10.1371/journal.pone.0062079
Characterization of the highly variable immune response gene family, He185/333, in the sea urchin, Heliocidaris erythrogramma
Abstract
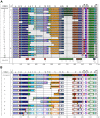
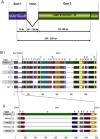
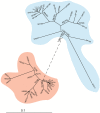
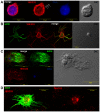
This study characterizes the highly variable He185/333 genes, transcripts and proteins in coelomocytes of the sea urchin, Heliocidaris erythrogramma. Originally discovered in the purple sea urchin, Strongylocentrotus purpuratus, the products of this gene family participate in the anti-pathogen defenses of the host animals. Full-length He185/333 genes and transcripts are identified. Complete open reading frames of He185/333 homologues are analyzed as to their element structure, single nucleotide polymorphisms, indels and sequence repeats and are subjected to diversification analyses. The sequence elements that compose He185/333 are different to those identified for Sp185/333. Differences between Sp185/333 and He185/333 genes are also evident in the complexity of the sequences of the introns. He185/333 proteins show a diverse range of molecular weights on Western blots. The observed sizes and pIs of the proteins differ from predicted values, suggesting post-translational modifications and oligomerization. Immunofluorescence microscopy shows that He185/333 proteins are mainly located on the surface of coelomocyte subpopulations. Our data demonstrate that He185/333 bears the same substantial characteristics as their S. purpuratus homologues. However, we also identify several unique characteristics of He185/333 (such as novel element patterns, sequence repeats, distribution of positively-selected codons and introns), suggesting species-specific adaptations. All sequences in this publication have been submitted to Genbank (accession numbers JQ780171-JQ780321) and are listed in table S1.
Conflict of interest statement
Figures



References
-
- Kurtz J (2005) Specific memory within innate immune systems. Trends in Immunology 26: 186–192. - PubMed
-
- Roth O, Kurtz J (2009) Phagocytosis mediates specificity in the immune defence of an invertebrate, the woodlouse Porcellio scaber (Crustacea: Isopoda). Developmental and Comparative Immunology 33: 1151–1155. - PubMed
-
- Kurtz J, Armitage S (2006) Alternative adaptive immunity in invertebrates. Trends in Immunology 27: 493–496. - PubMed
-
- Sadd BM, Schmid-Hempel P (2006) Insect immunity shows specificity in protection upon secondary pathogen exposure. Current Biology 16: 1206–1210. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

