Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage
- PMID: 25333463
- PMCID: PMC4409155
- DOI: 10.1038/ismej.2014.202
Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage
Abstract
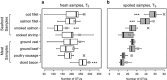
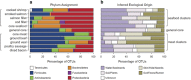
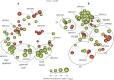
The microbial spoilage of meat and seafood products with short shelf lives is responsible for a significant amount of food waste. Food spoilage is a very heterogeneous process, involving the growth of various, poorly characterized bacterial communities. In this study, we conducted 16S ribosomal RNA gene pyrosequencing on 160 samples of fresh and spoiled foods to comparatively explore the bacterial communities associated with four meat products and four seafood products that are among the most consumed food items in Europe. We show that fresh products are contaminated in part by a microbiota similar to that found on the skin and in the gut of animals. However, this animal-derived microbiota was less prevalent and less abundant than a core microbiota, psychrotrophic in nature, mainly originated from the environment (water reservoirs). We clearly show that this core community found on meat and seafood products is the main reservoir of spoilage bacteria. We also show that storage conditions exert strong selective pressure on the initial microbiota: alpha diversity in fresh samples was 189±58 operational taxonomic units (OTUs) but dropped to 27±12 OTUs in spoiled samples. The OTU assemblage associated with spoilage was shaped by low storage temperatures, packaging and the nutritional value of the food matrix itself. These factors presumably act in tandem without any hierarchical pattern. Most notably, we were also able to identify putative new clades of dominant, previously undescribed bacteria occurring on spoiled seafood, a finding that emphasizes the importance of using culture-independent methods when studying food microbiota.
Figures
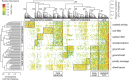

References
-
- Argyri AA, Doulgeraki AI, Blana VA, Panagou E, Nychas GJE. Potential of simple HPLC-based approach for the identification of the spoilage status of minced beef stored at various temperature and packaging systems. Int J Food Microbiol. 2001;150:25–33. - PubMed
-
- Baele M, Vancanneyt M, Devriese LA, Lefebvre K, Swings J, Haesebrouck F. Lactobacillus ingluviei sp. nov., isolated from the intestinal tract of pigeons. Int J Syst Evol Microbiol. 2003;53:133–136. - PubMed
-
- Banat IM, Marchant R, Rahman TJ. Geobacillus debilis sp. nov., a novel obligately thermophilic bacterium isolated from a cool soil environment, and reassignment of Bacillus pallidus to Geobacillus pallidus comb. nov. Int J Syst Evol Microbiol. 2004;54:2197–2201. - PubMed
-
- Barott KL, Rodriguez-Brito B, Janouskovec J, Marhaver KL, Smith JE, Keeling P, et al. Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ Microbiol. 2011;13:1192–1204. - PubMed
-
- Borch E, Kant-Muermans ML, Blixt Y. Bacterial spoilage of meat and cured meat products. Int J Food Microbiol. 1996;33:103–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

